
Biology and impact of lineage plasticity in ALK-positive NSCLC: a narrative review
Introduction
Background
Advanced non-small cell lung cancers (NSCLCs) harboring activating fusions in the anaplastic lymphoma kinase (ALK) are highly sensitive to treatment with ALK-targeted tyrosine kinase inhibitors (TKIs). Unfortunately, despite significant improvements in clinical outcomes, acquired resistance to ALK TKIs inevitably occurs (1-11).
Rationale and knowledge gap
Resistance is most often mediated by second-site mutations in the ALK driver fusion (2-4), resulting in a tumor with sustained dependence on ALK signaling for proliferation and survival. Such ALK-mediated resistance can be identified by DNA sequencing of tissue or plasma and can be overcome by later generations of ALK TKIs (4). However, a rarer and less well understood mechanism of resistance that occurs in only about 1.2% of TKI-resistant ALK-positive tumors is lineage transformation, most commonly as a shift in histology from adenocarcinoma to neuroendocrine or squamous histology (12). Patients whose resistant tumors have undergone lineage transformation represent a clinical subset for whom we have limited data to recommend optimal treatment regimens.
While ALK-positive tumors make up approximately 3–7% of NSCLC (13,14), tumors harboring other mutually exclusive, targetable driver oncogenes such as EGFR (epidermal growth factor receptor), RET (rearranged tyrosine kinase gene during transfection), and ROS1 (ROS proto-oncogene 1) collectively comprise approximately 20% of lung adenocarcinomas (15). With some variation in overall response rates, each of these genomic subtypes follow the same basic paradigm of initial response to targeted therapies followed inevitably by treatment resistance and disease progression. As with ALK-positive NSCLC, lineage transformation is a rare but recurrent mechanism of treatment resistance across genomic subtypes (12,16-19).
Objective
Here we present a review of what is known of the biology and clinical implications of lineage transformation in NSCLC, drawing parallels across oncogenic driver subtypes but with a focus on ALK-positive lung adenocarcinoma. We present this article in accordance with the Narrative Review reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-867/rc).
Methods
We performed a search of the published literature using PubMed and clinicaltrials.gov databases between October 1, 2022 and November 30, 2022 (Table 1). Search terms used included ALK, EGFR, NSCLC, histologic transformation, TKI resistance, small cell lung cancer (SCLC), squamous, large cell neuroendocrine carcinoma (LCNEC), and epithelialal-to-mesenchymal transition (EMT). We included only English-language publications reporting original research regarding randomized controlled trials, prospective or retrospective cohort studies, case reports or series, case-control studies, translational preclinical studies, and relevant review articles. Included articles were selected and assessed by both authors.
Table 1
| Items | Specification |
|---|---|
| Date of search | October 1, 2022 to November 30, 2022 |
| Databases and other sources searched | PubMed and clinicaltrials.gov |
| Search terms used | ALK, EGFR, NSCLC, histologic transformation, TKI resistance, small cell lung cancer (SCLC), squamous, large cell neuroendocrine carcinoma (LCNEC), and epithelial-to-mesenchymal transition (EMT) |
| Timeframe | From August 2007 until October 2022 |
| Inclusion and exclusion criteria | Inclusion criteria: |
| • English-language articles | |
| • Article types were randomized controlled trials, prospective or retrospective cohort studies, case reports or series, case-control studies, translational preclinical studies, and relevant review articles | |
| Exclusion criteria: | |
| • Article not published in English | |
| • Article types were editorial comments, abstracts, conference materials, review articles, guidelines, consensus statements, or study protocol | |
| Selection process | Study selection and full-text articles were assessed by CB Meador and Z Piotrowska |
ALK-positive NSCLC: clinical overview
Fusion rearrangements resulting in constitutive activation of the ALK tyrosine kinase domain were initially identified in NSCLC in 2007 and occur in approximately 3–7% of NSCLCs, most commonly in lung adenocarcinomas (13,14). EML4-ALK rearrangements represent the majority of oncogenic ALK fusions occurring in NSCLC; however, with application of more advanced next-generation sequencing platforms for tumor genotyping, approximately 90 total fusion partners for ALK have been identified in NSCLC to date (20). Activating ALK fusions in NSCLC are associated with adenocarcinoma histology, younger age and minimal smoking history as compared to NSCLCs that do not harbor ALK rearrangements (14).
After a decade of successful drug development in this area, there are five ALK TKIs now approved in the first-line setting as monotherapy for patients with stage IV ALK-positive lung adenocarcinoma (21-36). Crizotinib, a first generation ALK TKI, earned the first such approval in 2011 on the basis of overall response rates greater than 50% in early phase trials (21-23) and subsequent confirmatory phase III studies demonstrating superior outcomes compared to chemotherapy (24). The ability of investigators and regulatory entities to achieve clinical approval of crizotinib such a short time from discovery of ALK oncogenic fusions in NSCLC was due to the fact that crizotinib was initially developed as a MET (mesenchymal-epithelial transition factor) inhibitor but was subsequently found to have ALK inhibitory properties on kinase screens. For this reason it could be rapidly repurposed for clinical testing in ALK-positive NSCLC.
Second-generation ALK TKIs ceritinib and alectinib were FDA-approved for first-line treatment in 2017 and second-generation ALK TKI brigatinib was FDA-approved for first-line treatment in 2020 (1). Initially developed to overcome acquired resistance to crizotinib, these second-generation ALK TKIs proved to be more potent ALK inhibitors than crizotinib (2,37,38). Phase III trials comparing second-generation ALK TKIs head-to-head with crizotinib demonstrated a reproducible improvement in progression-free and overall survival as well as improved CNS outcomes for patients treated with second-generation ALK TKIs compared to crizotinib in the first-line setting (32-34).
Finally, lorlatinib is a third generation ALK TKI designed to be highly potent and selective in order to effectively overcome a range of resistance mutations known to be acquired following treatment with first- and second-generation ALK TKIs (35,36,39,40). Lorlatinib was initially FDA approved in 2018 for patients following treatment with alectinib or ceritinib (either as second line or after both crizotinib and second-generation ALK TKI) based on demonstrated response rate of 40% and median progression-free survival (mPFS) 6.9 months in this setting (41). FDA approval for lorlatinib in the first-line setting came in 2021 after publication of the phase III trial comparing first-line treatment with lorlatinib versus crizotinib demonstrated significantly greater overall response rate (76% vs. 58%) and higher 12-month progression-free survival (78% vs. 39%) in the lorlatinib treatment arm (42). Of note lorlatinib was intentionally designed to achieve excellent CNS penetration and the superior CNS efficacy was evident in this trial which demonstrated a significantly longer time to CNS disease progression (42).
Mechanisms of resistance to ALK TKIs
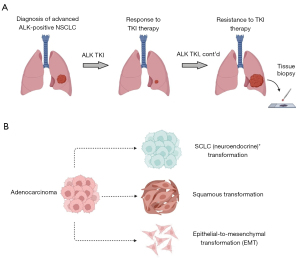
Despite the impressive clinical responses seen in metastatic ALK-positive NSCLC in response to ALK TKIs, unfortunately, disease progression inevitably occurs (Figure 1A). Repeat biopsies of tumor tissue (and, more recently, plasma) at the time of TKI resistance have provided insights into the mechanisms of resistance to ALK targeted therapies and their relative frequency across multiple generations of ALK TKIs (2,3,6). While a comprehensive review of mechanisms of resistance to ALK TKIs is beyond the scope of this narrative review, we outline below a broad overview of the field in order to put our discussion of lineage transformation in context.
Broadly speaking, resistance mechanisms can be categorized into ‘on-target’ and ‘off-target’ biological processes. ‘On-target’ mechanisms of resistance are defined as ALK-dependent, or ALK-mediated, resistance mechanisms and are the most common type of resistance mechanism that develops in the face of ALK TKI treatment with a variety of ALK resistance mutations described to date (2). In the case of crizotinib, ALK-dependent acquired resistance often occurs as acquired ALK L1196M mutations or via ALK amplification suggesting insufficient ALK inhibition with crizotinib therapy (2). In general, the landscape of acquired ‘on-target’ ALK resistance mutations varies by specific inhibitor, but for second-generation ALK TKIs the G1202R ALK solvent-front mutation is among the most commonly identified at the time of resistance, and compound ALK mutations also occur with some frequency following resistance to both second-generation and third-generation ALK TKIs (2,6). Following acquired resistance to lorlatinib, the frequency of ‘on-target’ resistance mutations decreases to 25–30% frequency (from >50% frequency following second-generation inhibitors) and preclinical data suggest that this figure may be evolve even lower with use of lorlatinib in the first-line setting (3,4).
‘Off-target’ mechanisms of resistance to ALK TKIs refer to acquired activation of ALK-independent pathways sustaining tumor growth in the setting of ongoing ALK inhibition. The most common form of ‘off-target’ resistance is bypass pathway signaling, including activation of either parallel receptor tyrosine kinase activation or downstream kinase signaling pathways. Bypass pathway resistance mechanisms identified at the time of crizotinib resistance include activation of signaling of receptor tyrosine kinases EGFR, HER2 (human epidermal growth factor receptor 2), KIT (proto-oncogene c-KIT) (43,44) and IGF-1R (insulin like growth factor 1 receptor) (45), downstream KRAS (kirsten rat sarcoma viral oncogene homolog) mutation (46), downstream YES1 (YES proto-oncogene 1) amplification (47), and DUSP6 (dual specificity phosphatase 6) loss (46). Bypass pathway activation reportedly conferring resistance to second- and third-generation ALK TKIs include MET amplification/mutation (48,49), RET rearrangement (50), HER2 amplification (51), and mutations or amplification in NF2 (neurofibromatosis type 2) (10), YES1 (47), BRAF (B-Raf Proto-Oncogene) (52), MAP2K1 (Mitogen-Activated Protein Kinase Kinase 1) (50,53), and PIK3CA (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha) (10).
Another form of ‘off-target’ resistance to ALK TKIs, and the primary focus of this narrative review, is lineage transformation. Originally reported as a mechanism of resistance to EGFR TKIs in EGFR-mutant NSCLC (16,18,54,55), lineage transformation has been identified as a recurrent driver-independent mechanism of resistance to targeted therapies across genomic subtypes of NSCLC and even among other solid tumor types (56). Because it is a rare phenomenon, the exact frequency of lineage transformation in the context of ALK-positive NSCLC has not been well established. Studies suggest a rate of at least 1.2% of SCLC transformation following next-generation ALK TKIs (12), and case reports demonstrate the occurrence of not only SCLC transformation but also squamous transformation and epithelial-to-mesenchymal transformation (EMT) following treatment with first, second and third-generation ALK TKIs (2,10,57-77) (Table 2 and Table 3).
Table 2
| Baseline histology | Trans histology | Age (years) | Sex | Smoking status | No. prior therapies | ALK TKI at trans | Time to trans (mos) | Co-occurring mutations or amplifications | Treatment post-trans | Response post-trans | OS (mos) | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adeno | SCLC | 41 | M | Never | 4 | Alectinib | 72 | Unk | Cisplatin + Irinotecan | PR | Unk | Yamagata (57) |
| Adeno | SCLC | 35 | F | Never | 2 | Crizotinib | Unknown | Unk | Ceritinib | PR | Unk | Yamagata (57) |
| Adeno | SCLC | 35 | M | Never | 2 | Lorlatinib | 10 | ALK G1202R | Carboplatin + Etoposide + Alectinib | PD | 11 | Ou (67) |
| Adeno | SCLC | 53 | F | Never | 2 | Ceritinib | 14 | PTEN T319del p53 V203M | Cisplatin + Etoposide | SD | Unk | Levacq (68) |
| Adeno | SCLC | 67 | F | Never | 7 | Alectinib | 27 | Unk | Irinotecan + Alectinib | PR | Unk | Fujita (69) |
| Adeno | SCLC | 63 | F | Never | 2 | Crizotinib | 6 | None | Alectinib | PR | Unk | Caumon (70) |
| Adeno | SCLC | 72 | M | Former (40 py) | 2 | Crizotinib | 6 | Not tested | Unk | Unk | Unk | Cha (71) |
| Adeno | SCLC | 43 | F | Unk | 8 | Alectinib | 41.5 | Not tested | None | N/A | Unk | Takegawa (72) |
| Adeno | SCLC | 56 | F | Never | 4 | Alectinib | 47 | Not tested | Cisplatin + Irinotecan | PD | Unk | Miyamoto (73) |
| Adeno | SCC | 49 | F | Former (5 py) | 3 | Brigatinib | 60 | MLH1 E37Q KMT2D L1461T KMT2D P647H NFE2L2 E82D |
Lorlatinib | Unk | Unk | Ball (59) |
| Adeno | SCC | 47 | F | Never | 3 | Ceritinib | 24 | None | Anlotinib | SD | Unk | Zhang (60) |
| Adeno | SCC | 53 | F | Never | 2 | Ceritinib | 4 | Not tested | Chemotherapy, NOS | PD | 6 | Kaiho (62) |
| Adeno | SCC | 60 | F | Former (4 py) | 3 | Alectinib | 43 | None | Pembrolizumab | PD | 48 | Gong (63) |
| Adeno | SCC | 52 | F | Never | 2 | Alectinib | 60 | Unk | Repotrectinib | PR | Unk | Park (65) |
| Adeno | SCC | 58 | F | Never | 9 | Lorlatinib | 138 | MET amp | Unk | Unk | Unk | Ueda (74) |
| Adeno | LCNEC | 74 | F | Never | 1 | Alectinib | 17 | ALK 1202R ALK V1180L | Brigatinib | PR | Unk | Fares (61) |
| Adeno | LCNEC | 54 | F | Never | 6 | Brigatinib | 75 | None | Carboplatin + Pemetrexed | PD | 81 | Fares (61) |
| 30% Adeno 70% LCNEC | N/A | 73 | F | Never | 0 | N/A | N/A | Not tested | Cisplatin + Pemetrexed + Alectinib | N/A (adjuvant) | 12 | Sim (66) |
| 10% Adeno 90% SCLC | N/A | 73 | M | Former (40 py) | 0 | N/A | N/A | Not tested | None | N/A (adjuvant) | 2 | Sim (66) |
| 10% Adeno 90% SCLC | N/A | 64 | M | Current (60 py) | 0 | N/A | N/A | Not tested | Cisplatin + Pemetrexed | N/A (adjuvant) | 18 | Sim (66) |
| Adeno | Focal SCLC | 36 | F | Never | 1 | Alectinib | 8 | Not tested | Cisplatin + Irinotecan | PR | Unk | Koyama (64) |
| Adeno | EMT | 36 | F | Never | 3 | Alectinib | 6.5 | MET amp p53 R248Q | Alectinib + abraxane + Cisplatin | PR | 19 | Xie (76) |
| Adeno | EMT | Unk | Unk | Unk | 4 | Ceritinib | >26 | ALK F1174C | None | N/A | Unk | Gainor (2) |
| Adeno | EMT | Unk | Unk | Unk | 2 | Ceritinib | 18 | None | Unknown (clinical trial) | Unknown | Unk | Gainor (2) |
| Adeno | EMT | Unk | Unk | Unk | 2 | Ceritinib | 14 | ALK L1196M | Chemotherapy, NOS | Unknown | Unk | Gainor (2) |
| Adeno | EMT | Unk | Unk | Unk | 5 | Ceritinib | >9 | ALK L1196M | None | N/A | Unk | Gainor (2) |
| Adeno | EMT | Unk | Unk | Unk | 2 | Ceritinib | 8 | None | Chemotherapy, NOS | Unk | Unk | Gainor (2) |
| Adeno | EMT | Unk | F | Unk | 1 | Crizotinib | 4 | ALK L1196M | Unk | Unk | 4 | Fukuda (75) |
| Adeno | EMT | 38 | M | Never | 4 | Alectinib | 36 | BRAF V600E | Lorlatinib | PD | 37 | Urbansk (77) |
| Adeno | EMT | 58 | F | Never | 3 | Lorlatinib | 64 | ALK C1156Y | Lorlatinib* | N/A | 96 | Recondo (10) |
*, Lorlatinib continued after cryoablation to transformed lesion. ALK, anaplastic lymphoma kinase; NSCLC, non-small cell lung cancer; TKI, tyrosine kinase inhibitor; Adeno, adenocarcinoma; SCLC, small cell lung cancer; SCC, squamous cell carcinoma; LCNEC, large cell neuroendocrine carcinoma; EMT, epithelial-to-mesenchymal transition; trans, transformation; Unk, unknown; amp, amplification; py, pack years; OS, overall survival; PR, partial response; PD, progressive disease; SD, stable disease; N/A, not available; NOS, not otherwise specified.
Table 3
| Transformed histology (references) | SCLC (57,67-74) | SCC (59,60,62,63,65) | LCNEC (61) | EMT (2,10,75-77) | Mixed histology (64,66) |
|---|---|---|---|---|---|
| Total number | 9 | 6 | 2 | 9 | 4 |
| Age at diagnosis if known (years)*, median [range] | 53 [35–72] | 52.5 [47–60] | 64 [54–74] | 38 [36–58] | 68.5 [36–73] |
| Smoking hx, n [%] | |||||
| Never | 7 [78] | 4 [67] | 2 [100] | 3 [33] | 2 [50] |
| Former; Avg pack years | 1 [11]; 40 | 2 [33]; 4.5 | – | – | 2 [50]; 50 |
| Unknown | 1 [11] | – | – | 6 [67] | – |
| Time to transformation in mos**, median [range] | 20.5 [6–72] | 51.5 [4–138] | 46 [17–75] | 14 [4-64] | 8 [8] |
| ALK TKI at the time of transformation^, n [%] | |||||
| Crizotinib | 3 [33] | – | – | 1 [11] | – |
| Alectinib | 4 [44] | 2 [33] | 1 [50] | 2 [22] | 1 [25] |
| Ceritinib | 1 [11] | 2 [33] | – | 5 [56] | – |
| Brigatinib | – | 1 [17] | 1 [50] | – | – |
| Lorlatinib | 1 [11] | 1 [17] | – | 1 [11] | – |
| Treatment after transformation, n [%] | |||||
| ALK TKI monotherapy | 2 [22] | 1 [17] | – | 2 [22] | – |
| Chemotherapy | 3 [33] | 1 [17] | 2 [100] | 2 [22] | 3 [75] |
| ALK TKI + chemotherapy | 2 [22] | – | – | 1 [11] | – |
| Other/unknown | 2 [22] | 4 [67] | – | 4 [44] | 1 [25] |
| Response after transformation, n [%] | |||||
| PR | 4 [44] | 1 [17] | 1 [50] | 1 [11] | 1 [25] |
| SD | 1 [11] | 1 [17] | – | – | – |
| PD | 2 [22] | 2 [33] | 1 [50] | 1 [11] | – |
| Unknown | 2 [22] | 2 [33] | – | 7 [78] | 3 [75] |
*, 5 pts omitted as age not reported; **, 6 pts omitted as time to transformation not reported or not applicable (mixed histology at diagnosis); ^, 3 pts omitted as they were not treated prior to mixed histology biopsy. ALK, anaplastic lymphoma kinase; NSCLC, non-small cell lung cancer; Adeno, adenocarcinoma; SCLC, small cell lung cancer; SCC, squamous cell carcinoma; LCNEC, large cell neuroendocrine carcinoma; EMT, epithelial-to-mesenchymal transition; TKI, tyrosine kinase inhibitor; PR, partial response; PD, progressive disease; SD, stable disease.
A comprehensive understanding of the biology of lineage transformation in ALK-positive NSCLC is limited in part by its infrequent occurrence and lack of robust patient-derived tissue samples for study. In addition, the complexity of factors governing cellular differentiation make this an especially challenging biologic process to define. However, lineage state is clearly a critical component of our understanding of tumor initiation and evolution across the spectrum of malignancies. As alluded to above, within the scope of the lung cancer literature, insights into the biology of lineage transformation can be broadly grouped into three categories for discussion: SCLC histologic transformation, squamous histologic transformation, and EMT (Figure 1B).
SCLC histologic transformation
Incidence and diagnosis of SCLC transformation
Transformation from NSCLC to SCLC histology was initially reported as a mechanism of TKI resistance in EGFR-mutant NSCLC (54). As a result, much of our knowledge of the biology, risk factors, and clinical outcomes of SCLC transformation comes from the EGFR-mutant NSCLC literature. In initial studies of resistance mechanisms to early-generation EGFR TKIs that collectively included 192 patients, SCLC transformation was identified in 3–14% of TKI-resistant tumors (54,78). The incidence of SCLC transformation at the time of resistance to third-generation EGFR TKI osimertinib is estimated at 5–7% based on recent studies that included a total of 103 patients, though these estimates are limited by the fact that many of the largest series of osimertinib resistance have not evaluated tumor histology at the time of progression (17,19,79,80). There are also multiple case reports and case series demonstrating SCLC transformation following EGFR TKIs (58,81-95).
By comparison, available data suggest that SCLC transformation occurs less frequently in ALK-positive NSCLC than in EGFR-mutant NSCLC. The largest published studies analyzing repeat biopsies after ALK TKI resistance did not identify any cases of SCLC among 91 patients with resistance to crizotinib (2,12), a 0–0.08% frequency (out of 157 total patients) after resistance to second-generation ALK TKIs (2,12), and a 2.7% (out of 38 total patients) frequency of SCLC transformation after resistance to lorlatinib (12). Of the 9 published case reports of SCLC transformation as a mechanism of ALK TKI resistance, 3 occurred following crizotinib, 5 occurred following second-generation TKIs, and 1 occurred following lorlatinib (Table 2). Notably, while it is seen most commonly following resistance to EGFR and ALK targeted therapies, SCLC histologic transformation has been reported at the time of acquired resistance to targeted therapy in a NSCLC harboring a driver ROS1 fusion (12) as well as following treatment with immune checkpoint inhibitors in NSCLC negative for oncogenic driver alterations (96-99). The relative incidence of SCLC transformation following different therapies, across genotypes and clinical presentation of NSCLC remains an area of ongoing study.
SCLC transformation is diagnosed by histopathologic examination of tissue biopsy at the time of acquired resistance to therapy. Morphologically and immunohistochemically, transformed SCLC mimics the appearance of de novo SCLC (classic, non-transformed SCLC), characterized by small cells with high nuclear-to-cytoplasmic ratio, high mitotic activity and positive staining for neuroendocrine markers such as chromogranin and synaptophysin (12,18,54,100). Genomically, the majority of transformed SCLC tumors harbor loss-of-function mutations in tumor suppressors TP53 (tumor protein 53) and RB1 (RB transcriptional corepressor 1), which is also characteristic of de novo SCLC tumors (12,18,54,100). In fact, an EGFR-mutant NSCLC with complete inactivation of p53 and RB1 at baseline is 43 times more likely to evolve to SCLC as a mechanism of resistance to TKI therapy compared to an EGFR-mutant NSCLC without these co-occurring alterations at baseline (101). Finally, while the original EGFR driver mutation is genomically preserved in the SCLC transformed tumor, loss of expression of EGFR is seen at the protein level suggesting loss of dependence on EGFR oncogenic signaling for these transformed tumors (16,18,100).
In published cases of SCLC transformation of ALK-positive NSCLC, transformed SCLC is similarly reported to have classic SCLC morphology with acquired expression of neuroendocrine markers; however, expression of ALK protein is variable and not uniformly lost as with EGFR-mutant SCLC transformed tumors (57,67-73). Of the small overall number of published cases of SCLC transformation in ALK-positive NSCLC, only a handful report baseline next-generation panel-based sequencing data. As a result, there are not yet clearly established trends regarding rates of co-occurring mutations, for example loss-of-function mutations in p53 and/or RB1.
Of note, another histologic entity that falls under the spectrum of neuroendocrine histology but is distinct from SCLC is large cell neuroendocrine carcinoma (LCNEC). LCNEC is characterized by cytologic features of NSCLC with tissue architecture and IHC markers consistent with neuroendocrine histology (102). The largest published cohort of genomic and transcriptomic profiling of LCNEC tumors to date revealed two distinct genomic subgroups defined by bi-allelic TP53 and STK11 (serine/threonine kinase 11)/KEAP1 (Kelch Like ECH Associated Protein 1) alterations (type I) versus bi-allelic inactivating mutations in TP53 and RB1 (type II) (103). While LCNEC tumors are genomically similar to NSCLC, transcriptional profiling demonstrates that type I LCNEC resembles neuroendocrine-high SCLC, and type II LCNEC more closely resembles neuroendocrine-low SCLC (103). A full characterization of LNEC is outside the scope of this review, but the significant degree of histologic heterogeneity within LCNEC highlights the inherent lineage plasticity of this tumor subtype and suggests that improved understanding of the molecular features of LCNEC will lead to more precision in diagnosis and treatment decision-making.
While these descriptions refer to de novo LCNEC tumors, histologic transformation from adenocarcinoma to LCNEC is also rarely identified as a mechanism of resistance to targeted therapies, including ALK TKIs (61,83,104-106) (Table 2). Molecular features associated with LCNEC transformation remain poorly defined, but there is variability in baseline p53/RB1 status and clinical outcomes among the few reported cases. Within EGFR-mutant NSCLC, there are reported cases of both TKI-sensitive and -resistant de novo EGFR-mutant LCNEC tumors (107-109), as well as published examples of histologic transformation to LCNEC following treatment with EGFR-targeted therapies (83,106,110). Though the published examples of such cases are limited in number, the observed variability in responses to EGFR TKIs among EGFR-mutant tumors with LCNEC histology likely reflects the inherent molecular and histologic heterogeneity of LCNEC tumors more broadly.
Molecular features of SCLC transformation
As mentioned in the prior section, loss of function of tumor suppressors p53 and RB1 appears to be a critical biologic factor predisposing tumors to undergo SCLC transformation. The combined loss of these p53/RB1 function has been described as necessary but not sufficient for SCLC transformation in the context of EGFR-mutant NSCLC (101). This is consistent with the genomic landscape of de novo SCLC in which >95% of tumors are deficient in both p53/RB1 (111-113). Beyond p53/RB1 loss, however, multiple additional molecular mechanisms have been proposed as potentially important pathways in SCLC transformation. In the largest retrospective clinical cohort of transformed EGFR-mutant SCLC published to date, PIK3CA mutations were reported in 27% (n=14/52) of pre-transformed tumors (18) which was consistent with recurrent PIK3CA mutations observed in the initial report of SCLC transformation (54) and other subsequent case series (88). In another study comparing 7 pre-SCLC transformation tumors to 32 NSCLC tumors with baseline EGFR, TP53, and RB1 mutations that did not go on to SCLC transformation, the pre-transformed tumors were enriched for mutations or amplifications in SMYD1 (SET and MYND domain containing 1), MYND (Mynd Domain-Containing Protein), NOTCH2 (neurogenic locus notch homolog protein 2), PIK3CA, MYC (Cellular Myelocytomatosis Oncogene), CREBBP (CREB-binding protein), PTEN (Phosphatase and TENsin homolog), CCNE1 (Cyclin E1) and ELF3 (E74 like ETS transcription factor 3) (16). Pre-transformed tumors were also enriched for whole-genome duplication (WGD) compared to EGFR/TP53/RB1 co-mutated tumors that did not transform (16), and multiple studies have also identified APOBEC (Apolipoprotein B mRNA-Editing Enzyme, Catalytic Polypeptide) hypermutation signatures in pre-transformed EGFR-mutant NSCLC (16,101). Other case reports and case series of transformed SCLC tumors suggest that increased expression of FGF9 (fibroblast growth factor 9) (114), amplification of TERT (telomerase reverse transcriptase) (81), and overall increased burden of copy-number variants (115) may also play a role in SCLC transformation.
Mixed histology tumors have also become a valuable resource for comparing the biology of discordant histologic components of a single biopsy or resection specimen. In a series of 100 surgically resected cases of NSCLC, a component of SCLC or LCNEC histology was seen in 25% of cases (116). A recent report describing 11 EGFR-mutant SCLC transformed tumors from a retrospective cohort of 7282 cases of lung cancer demonstrated discernable foci of SCLC histology in 8/11 ‘pre-transformation’ pathology specimens suggesting a true diagnosis of mixed histology at baseline (84). Finally, some component of mixed histology was present at the time of transformation in a subset of reported cases of EGFR-mutant NSCLC (54,92) and ROS1-rearranged transformed SCLC (12).
With increasingly powerful investigative tools such as spatial transcriptomics and single-cell sequencing technologies, we continue to gain more insights into the biology of these mixed histology tumors. A recent study comparing genomic, transcriptomic, and methylomic data from 11 mixed histology (adenocarcinoma/SCLC) tumors, 5 pre-transformed lung adenocarcinomas, and 3 post-transformed SCLCs (including one matched case) to never-transformed lung adenocarcinomas (n=15) and to de novo SCLCs (n=18) was arguably the most comprehensive analysis of transformed and mixed NSCLC/SCLC cases published to date (117). In it, the authors demonstrate consistent genomic findings in NSCLC and SCLC components as previously published; specifically, enrichment of both chromosomal 3p loss and APOBEC hypermutation signatures is demonstrated in the NSCLC portions of mixed histology tumors, and increased expression of genes related to the PRC2 (polycomb repressive complex 2) complex, PI3K/AKT (Protein Kinase B) signaling, and NOTCH signaling pathways is seen in SCLC components of transformed or mixed histology tumors (117). Interestingly but not surprisingly, transcriptional and methylation data reveal substantial overlap between the respective NSCLC and SCLC components of mixed histology tumors as compared to their never-transformed comparison cohorts (117). This finding affirms the hypothesis that these pre-/post-transformed and mixed histology tumors represent an intermediate state of lineage plasticity and further strengthens the view that the process of histologic transformation is likely one governed by transcriptional reprogramming. However, while there has been work defining key transcriptional programs governing SCLC tumorigenesis (118), the mechanisms driving gene expression and methylation changes in the process of SCLC transformation remain incompletely understood.
While it’s important to note that directionality of histologic transformation cannot be assumed in a mixed histology tumor, the available published data summarized above demonstrate clonality within mixed histology tumors. The presence of mixed histologic components prior to TKI or other treatment exposure raises the question of whether (or when) driver oncogene inhibition is required for the process of SCLC histologic transformation. While loss of EGFR expression in SCLC-transformed EGFR-mutant NSCLCs demonstrates a repression of driver oncogene pathway activity in SCLC transformed tumors (16,18,54), multiple studies investigating the clonal evolution of SCLC transformation suggest that, in some cases, the divergence of precursor SCLC clones occurs prior to TKI exposure (101,115).
Finally, while the vast majority of de novo SCLC tumors show RB1 deficiency, the ~5% of SCLC tumors that are RB1 proficient appear to represent a distinct subset characterized by decreased expression of classical neuroendocrine markers, distinct genomic alterations [CDKN2A (cyclin-dependent kinase inhibitor 2A) mutation, CCND1 (cyclin D1) amplification, KEAP1/STK11 mutations, FGFR1 (fibroblast growth factor receptor 1) amplification] and a more aggressive clinical phenotype (113). These RB1 proficient tumors also more commonly demonstrate features of SCLC/NSCLC mixed histology, suggesting that ‘mixed histology’ and ‘neuroendocrine-low’/RB1-proficient SCLC tumors may describe some component of an overlapping histologic subtype. In the context of clinically observed histologic transformations from adenocarcinoma to SCLC histology, extrapolation of data from mixed histology tumors is, at a minimum, hypothesis-generating, but may also provide key insights into mechanisms of lineage plasticity. Taken together, the sum of the data from the EGFR-mutant NSCLC literature about SCLC transformation suggests that it is a biologic process driven primarily by transcriptional reprogramming rather than acquired genomic alterations.
Treatment approaches and clinical outcomes following SCLC transformation
As a field, we lack prospective data informing clinical treatment of patients with SCLC transformed tumors. The largest retrospective cohort published to date includes data from 58 patients with EGFR-mutant NSCLC across 8 institutions whose tumors underwent SCLC transformation as a mechanism of EGFR TKI resistance (18). In it, the authors demonstrated an overall response rate of 54% and mPFS of 3.4 months after treatment with platinum/etoposide-based chemotherapy. Taxane-containing chemotherapy regimens also conferred promising clinical benefit with a 50% overall response rate and mPFS of 2.7 months. No responses to immune checkpoint inhibitors were seen in this cohort. The median time to SCLC transformation was 17.8 months and the mOS was 31.5 months. Median OS from the time of SCLC transformation was 10.9 months, comparable to the expected survival of de novo SCLC from the time of initial diagnosis (119,120).
Notably, the response rate to platinum etoposide chemotherapy reported in this retrospective case series is somewhat lower than the historical average of 60–65% to first-line platinum/etoposide-based chemotherapy in de novo SCLC (121,122). Though direct comparisons cannot be drawn between this retrospective study and independent prospective clinical trials, these data suggest decreased sensitivity to platinum-based chemotherapy regimens in the setting of transformed SCLCs compared to de novo SCLCs. Interestingly, the response rate to platinum/etoposide in patients who were previously treated with platinum-based chemotherapy for pre-transformed adenocarcinoma was 80% (n=8/10) suggesting that, if the lower overall response rate reflects truly differing clinical rates of response, this is more likely due to the underlying biology of transformed SCLCs than a result of prior platinum-based chemotherapy exposure (18). A more recently published cohort of 29 patients with EGFR-mutant NSCLC whose tumors underwent SCLC transformation following treatment with EGFR TKIs demonstrated a median time to transformation of 27.5 months and mOS from the time of SCLC transformation of 14.8 months (82). The mPFS for patients who received EGFR TKI treatment concurrently with chemotherapy at the time of SCLC transformation was significantly longer than those who received chemotherapy alone (5.0 vs. 3.7 mos) but there was no significant difference between the two groups in mOS from the time of transformation.
No such large retrospective cohort analyses exist in the published literature for ALK-positive NSCLC following SCLC transformation, but data compiled from case series suggest an average time to SCLC transformation of 20.5 months (Table 3). The most commonly reported treatment regimens following SCLC transformation in ALK-positive lung cancer are platinum-based chemotherapy alone with or without ALK TKI (Table 2). Response data from case reports of SCLC transformed ALK-positive lung cancers following specific treatment regimens are detailed in Table 2. Of note, there is at least one reported case of successful retreatment with alectinib after eradication of a SCLC histologic clone following transformation (57).
While no prospective trial data are yet available to guide treatment of SCLC transformed tumors, the first generation of clinical trials designed for this specific patient population are open to enrollment (Table 4). To our knowledge, all three of these trials enrolling in the United States are open only to EGFR-mutant transformed SCLC, not ALK-positive transformed SCLC or other non-EGFR-mutant subsets. Extrapolating from the de novo SCLC literature (123), two trials are testing combination therapies with PD-L1 (programmed death-ligand 1) inhibitor durvalumab at the time of SCLC transformation, either together with PARP (Poly-ADP Ribose Polymerase 1) inhibitor olaparib (NCT04538378) or with platinum/etoposide chemotherapy (NCT03944772; arm of the ORCHARD platform study). Interestingly, the third study open for this patient population is designed with the aim of delaying or preventing SCLC transformation by eradicating pre-existing SCLC subclones prior to the clinical emergence of transformation. This study enrolls patients with newly-diagnosed, high-risk EGFR-mutant NSCLC (those with concurrent TP53/RB1 mutations at baseline) and prospectively adds four cycles of platinum/etoposide chemotherapy prior to transformation, after 12 weeks of standard-of-care osimertinib monotherapy (NCT03567642).
Table 4
| NCT identifier | Trial phase | Patient population | Intervention | Primary outcome(s) | Trial status |
|---|---|---|---|---|---|
| NCT04538378 | Phase II | Stage IV EGFR-mutant NSCLC after transformation to SCLC or other NEC | Olaparib 300 mg bid plus durvalumab 1,500 mg monthly | Objective response rate | Recruiting |
| NCT03567642 | Phase I | Stage IV EGFR-mutant NSCLC with co-occurring baseline TP53 and RB1 mutations | Addition of cisplatin (60 mg/m2) or carboplatin (AUC 4–5) with etoposide (80–100 mg/m2) starting C4D1 osimertinib 80 mg daily | Maximum tolerated dose (MTD) | Recruiting |
| NCT03944772 | Phase II (Platform study) | Stage IV EGFR-mutant NSCLC after transformation to SCLC | Etoposide 80–100 mg/m2 plus durvalumab 1,500 mg plus either cisplatin or carboplatin | Objective response rate | Recruiting |
NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; NEC, neuroendocrine carcinoma.
Of note, the list of trials described here does not comprehensively describe all clinical trial opportunities for these patients, as other trials designed specifically for de novo SCLC or other neuroendocrine cancers may allow transformed SCLC on a case-by-case basis. Thinking forward to additional therapeutics in this space, there are emerging data demonstrating rationale for other targeted therapies of potential biologic important in transformed SCLC. An example of one potential molecular target is the EZH2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit) H3K27 methyltransferase, a catalytic subunit of the PRC2 complex which has been shown to be dysregulated in neuroendocrine transformation (117). Preclinical data have shown that inhibition of EZH2 can shift cells from neuroendocrine to non-neuroendocrine morphology and may play a role in SCLC lineage plasticity in NSCLC and other solid tumors (56,124). EZH2 inhibitors are currently being tested in early-phase clinical trials.
Finally, recent progress in translational studies of de novo SCLC has resulted in a novel classification system categorizing de novo SCLC into molecular subtypes based on gene expression of key transcription factors [ASCL1 (Achaete-Scute Homolog 1), NEUROD1 (neuronal differentiation 1), POU2F3 (POU Domain, Class 2, Transcription factor 3)] and immune-related genes (inflamed subtype). These classifications (SCLC-A, SCLC-N, SCLC-P, and SCLC-I) have been further shown in preclinical and retrospective studies to confer sensitivity to specific targeted therapies such as PARP inhibitors, AURK inhibitors, and BCL2 inhibitors (125); however they are not yet prognostic or predictive for patient outcomes. Initial data suggest representation of all transcription factor subtypes in transformed SCLC (117), though more investigation is required to understand the clinical relevance, if any, these subtypes will have in transformed SCLC.
Squamous histologic transformation
Incidence and diagnosis of squamous transformation
While SCLC transformation was the first identified category of lineage transformation and is the most comprehensively described to date, squamous transformation (from adenocarcinoma, within the NSCLC spectrum) is also a recurrent finding at the time of acquired resistance to targeted therapies in lung cancer. Characterized by changes in morphology and immunohistochemical staining from adenocarcinoma-associated markers (e.g., napsin A and TTF-1) to squamous-associated markers (e.g., p40 and p63) (126,127), it has been reported in at least six cases of resistance to ALK TKIs (59,60,62,63,85,74) (Table 2 and Table 3). Similar to SCLC transformation, squamous transformation is also a recurrent phenomenon in EGFR-mutant NSCLC (17,19,128,129) and squamous histology has also been rarely reported as a component of EGFR-mutant NSCLC histology at diagnosis (130,131). ALK fusions are an exceedingly rare occurrence in de novo squamous cell carcinoma tumors (132). Overall, the full clinical spectrum of squamous transformation from adenocarcinoma histology remains understudied, though it seems to represent a true recurrent adaptive mechanism for tumor evasion of targeted therapy.
Molecular features of squamous transformation
Similar to SCLC transformation, investigation of mixed adenosquamous tumors provides a compelling opportunity to learn about mechanisms of lineage transformation along the adenocarcinoma-squamous spectrum of NSCLC histology. Early studies performing molecular characterization of separate histologic compartments of surgically resected, macro-dissected mixed adenosquamous tumors demonstrated expected clonality between the adenocarcinoma and squamous tumor components (133). Specifically, whole-exome sequencing of three mixed adenosquamous tumors showed common genomic alterations in the identified driver mutation (EGFR, BRAF, and MET respectively) as well as shared loss of tumor suppressor STK11 and chromosomal regions 3p, 15q, 19p. In addition to chromosomal amplification of 5p, focal amplification of SOX2 (SRY-Box Transcription Factor 2) was also identified in all three tumors. SOX2 amplification was previously shown to cause squamous differentiation in preclinical mouse models (134,135) and is amplified in 40% of squamous cell carcinomas but almost never in lung adenocarcinomas (126).
In a recent study published by Quintanal-Villalonga and colleagues, the authors performed genomic, epigenomic, and transcriptomic analysis of 11 micro-dissected adenosquamous tumors, 4 pre-transformation adenocarcinoma tumors, and 7 post-transformation squamous tumors compared to never-transformed lung adenocarcinomas and de novo lung squamous cell carcinomas (136). Common mutations present in both adenocarcinoma and squamous histologic compartments were in EGFR, TP53, CDKN2A/B, and STK11, but these alterations were not specifically enriched in the transformed or mixed histology tumors compared to controls (136). However, mutations in TBX3 (T-Box transcription factor 3), MET, RBM10 (RNA Binding Motif Protein 10) were enriched in pre-transformed lung adenocarcinomas compared to never-transformed lung adenocarcinomas, and increased expression of genes related to PI3K/AKT, MYC, and PRC2 signaling pathways was found in squamous transformed components of either mixed histology or post-transformation tumors (136). While further investigation is needed to confirm the role of these signaling pathways in squamous transformation, these data raise the hypotheses that squamous transformation may be primed by genomic alterations in TBX3, MET, RBM10 and that the process of lineage transformation is driven by transcriptional reprogramming rather than further acquired genomic mutational changes.
Treatment approaches and clinical outcomes following squamous transformation
There are unfortunately very little clinical data informing optimal treatment regimens for patients whose tumors undergo squamous transformation following resistance to targeted therapies. Without any published prospective data or clinical trials specifically designed for this patient population, decisions regarding treatment at the time of squamous transformation still rely on anecdotal evidence and expert opinion. In reported cases of ALK-positive and EGFR-mutant tumors undergoing squamous transformation, systemic therapy at the time of transformation has been variable—ranging from continued TKI to chemotherapy and immune checkpoint inhibitors (Table 2) (17,59,60,62,63,65,74,137). Unfortunately, the number of cases with reported clinical outcomes remains too few to inform prospective decision-making for patients currently on treatment. In general, similar principles apply as in SCLC transformation, including a shift to histology-based chemotherapy backbones (e.g., taxane-based regimens for squamous histology) and continuation of concurrent targeted therapy if safe and feasible versus shift to alternative targeted therapy if concurrent activation of bypass pathways are found on molecular analysis. Of note, while squamous transformation has been purported to be a primary and/or independent driver of TKI resistance, retrospective analyses in EGFR-mutant tumors with squamous transformation have shown concurrent known genomic mechanisms of resistance (17,137). It is not yet known whether these findings represent intra-tumoral heterogeneity in resistance mechanisms versus biologic association of these genomic changes with squamous transformation.
In the case of NSCLCs with baseline mixed adenosquamous histology, historical data suggest that clinical outcomes are overall poorer compared to baseline pure adenocarcinoma (138). However, the genomic context of a particular oncogenic driver alteration and associated available treatment regimens is likely to be important for interpretation of prognosis and expected clinical outcomes. In EGFR-mutant NSCLCs, some component of admixed squamous histology at diagnosis was reportedly associated with comparable clinical outcomes in a retrospective study of 12 patients treated with first-generation EGFR TKI erlotinib (130). However, other preliminary data from a retrospective analysis of EGFR adenosquamous or purely squamous NSCLC at diagnosis suggests that these patients have inferior clinical outcomes compared to those with adenocarcinoma histology (137). We lack comparable data regarding clinical outcomes of ALK-positive mixed adenosquamous NSCLCs.
Epithelial-to-mesenchymal transition (EMT)
EMT is another example of lineage plasticity that has been observed in ALK-positive lung adenocarcinoma following resistance to targeted therapies (2,10,75-77). Less diagnostically definitive than a complete SCLC or squamous histologic transformation, EMT is characterized by a morphologic and immunophenotypic shift of increased expression of mesenchymal markers (e.g., vimentin) and decreased expression of epithelial markers (e.g., e-cadherin) (2). Multiple rigorous preclinical studies have further defined subtypes or distinct ‘modes’ of EMT and signaling pathways thought to be primarily responsible for driving this form of cellular plasticity across solid tumor types (139-141). While a comprehensive summary of these preclinically-defined mechanisms of EMT is outside the scope of this narrative review, it is important to note that the biology of EMT—including predisposing factors, definitive diagnostic criteria, and clinical implications—remains an area of active study. Of note, characterization of the neuroendocrine-low morphologic subvariant of SCLC has revealed some molecular and phenotypic similarities to EMT (142), but the molecular relationship between EMT and SCLC transformation remains incompletely understood. EMT has been reported as a mechanism of resistance in at least 9 cases of acquired resistance to ALK TKIs in ALK-positive NSCLC (Table 2 and Table 3) (2,10,75-77), though this is not a mechanism of resistance routine tested for or considered actionable on a clinical basis at this time.
Limitations and future directions
Lineage plasticity is an increasingly recognized but relatively rare mechanism of resistance to targeted therapies in ALK-positive NSCLC. Multiple forms of lineage plasticity have been implicated as methods of tumor cell evasion from the selective pressure of targeted therapies, and SCLC transformation, squamous transformation and EMT are the most well studied. This narrative review of the literature on lineage plasticity in ALK-positive NSCLC is limited in part by minimal available published data due to the rare occurrence of lineage transformation relative to other mechanisms of resistance to ALK TKIs. In order to address this limitation, we have referenced the relevant available literature about transformation in other genomic subtypes of NSCLC, such as EGFR-mutant NSCLC.
Distinct from genomic mechanisms of TKI resistance such as second-site oncogene mutations and bypass pathway amplification, lineage transformation does not have reliable genomic markers that can be assessed via DNA sequencing. This precludes diagnosis of lineage transformation by non-invasive plasma circulating tumor DNA sequencing assays, which are now commonly used to diagnose resistance mechanisms (143). As a result, lineage transformation can be easily missed if a repeat tissue biopsy is not obtained at the time of TKI resistance for the purpose of histologic analysis. At present, these tissue samples are also needed for ongoing translational studies to further understand the biology of lineage plasticity in NSCLC. In the future, non-invasive plasma-based diagnostic assays that could detect lineage plasticity would obviate the need for repeat tissue biopsies, but this technology is not yet readily available for clinical application.
We also still lack reliable predictive biomarkers identifying tumors predisposed to undergo lineage transformation. While dual TP53/RB1 loss portends a higher likelihood of SCLC transformation in EGFR-mutant lung adenocarcinoma, we have yet to define any other biomarker of a pre-transformed tumor that could potentially be clinically actionable. Discovery of predictive markers of lineage transformation would potentially enable more adaptive trial designs aimed at preventing or delaying lineage transformation, hopefully improving patient outcomes. Finally, the role that timing and potency of oncogene inhibition plays in driving lineage transformation also remains largely unknown. Whether TKI inhibition of driver oncogene (e.g., shutting down oncogenic epithelial signaling pathways) is required for lineage transformation in pre-disposed tumors and/or whether increasingly specific and potent TKIs will drive lineage transformation at higher frequencies compared to on-target resistance mechanisms remains unknown. Ongoing clinical and translational investigative efforts are needed to answer these questions more definitively and move clinical care forward for these patients.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Jessica J. Lin and Justin F. Gainor) for the series “ALK Positive NSCLC” published in Translational Lung Cancer Research. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-867/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-867/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-867/coif). The series "ALK Positive NSCLC” was commissioned by the editorial office without any funding or sponsorship. ZP reports consulting fees/honoraria for advisory board participation from Sanofi, Taiho, Janssen, Takeda, AstraZeneca, Cullinan Oncology, C4 Therapuetics, Jazz Pharmaceuticals and Blueprint, honoraria for educational events from Daiichi Sankyo, Janssen and Eli Lilly and research support (to institution) from Novartis, Takeda, Spectrum, AstraZeneca, Tesaro/GSK, Cullinan Oncology, Daiichi Sankyo, AbbVie, Janssen and Blueprint. The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cooper AJ, Sequist LV, Lin JJ. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat Rev Clin Oncol 2022;19:499-514. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer Discov 2018;8:714-29. [Crossref] [PubMed]
- Shiba-Ishii A, Johnson TW, Dagogo-Jack I, et al. Analysis of lorlatinib analogs reveals a roadmap for targeting diverse compound resistance mutations in ALK-positive lung cancer. Nat Cancer 2022;3:710-22. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney M, Lin JJ, et al. Treatment with Next-Generation ALK Inhibitors Fuels Plasma ALK Mutation Diversity. Clin Cancer Res 2019;25:6662-70. [Crossref] [PubMed]
- Pailler E, Faugeroux V, Oulhen M, et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer. Clin Cancer Res 2019;25:6671-82. [Crossref] [PubMed]
- Zhu VW, Nagasaka M, Madison R, et al. A Novel Sequentially Evolved EML4-ALK Variant 3 G1202R/S1206Y Double Mutation In Cis Confers Resistance to Lorlatinib: A Brief Report and Literature Review. JTO Clin Res Rep 2020;2:100116. [Crossref] [PubMed]
- Takahashi K, Seto Y, Okada K, et al. Overcoming resistance by ALK compound mutation (I1171S + G1269A) after sequential treatment of multiple ALK inhibitors in non-small cell lung cancer. Thorac Cancer 2020;11:581-7. [Crossref] [PubMed]
- Recondo G, Mezquita L, Facchinetti F, et al. Diverse Resistance Mechanisms to the Third-Generation ALK Inhibitor Lorlatinib in ALK-Rearranged Lung Cancer. Clin Cancer Res 2020;26:242-55. [Crossref] [PubMed]
- Pan Y, Deng C, Qiu Z, et al. The Resistance Mechanisms and Treatment Strategies for ALK-Rearranged Non-Small Cell Lung Cancer. Front Oncol 2021;11:713530. [Crossref] [PubMed]
- Lin JJ, Langenbucher A, Gupta P, et al. Small cell transformation of ROS1 fusion-positive lung cancer resistant to ROS1 inhibition. NPJ Precis Oncol 2020;4:21. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [Crossref] [PubMed]
- Meador CB, Sequist LV, Piotrowska Z. Targeting EGFR Exon 20 Insertions in Non-Small Cell Lung Cancer: Recent Advances and Clinical Updates. Cancer Discov 2021;11:2145-57. [Crossref] [PubMed]
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res 2020;26:2654-63. [Crossref] [PubMed]
- Marcoux N, Gettinger SN, O'Kane G, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol 2019;37:278-85. [Crossref] [PubMed]
- Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529-39. [Crossref] [PubMed]
- Ou SI, Zhu VW, Nagasaka M. Catalog of 5' Fusion Partners in ALK-positive NSCLC Circa 2020. JTO Clin Res Rep 2020;1:100015. [Crossref] [PubMed]
- Zou HY, Li Q, Lee JH, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res 2007;67:4408-17. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-9. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Kim DW, Mehra R, Tan DSW, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 2016;17:452-63. [Crossref] [PubMed]
- Ou SH, Ahn JS, De Petris L, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol 2016;34:661-8. [Crossref] [PubMed]
- Shaw AT, Gandhi L, Gadgeel S, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol 2016;17:234-42. [Crossref] [PubMed]
- Gettinger SN, Bazhenova LA, Langer CJ, et al. Activity and safety of brigatinib in ALK-rearranged non-small-cell lung cancer and other malignancies: a single-arm, open-label, phase 1/2 trial. Lancet Oncol 2016;17:1683-96. [Crossref] [PubMed]
- Kim DW, Tiseo M, Ahn MJ, et al. Brigatinib in Patients With Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase II Trial. J Clin Oncol 2017;35:2490-8. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib Versus Crizotinib in ALK Inhibitor-Naive Advanced ALK-Positive NSCLC: Final Results of Phase 3 ALTA-1L Trial. J Thorac Oncol 2021;16:2091-108. [Crossref] [PubMed]
- Horn L, Wang Z, Wu G, et al. Ensartinib vs Crizotinib for Patients With Anaplastic Lymphoma Kinase-Positive Non-Small Cell Lung Cancer: A Randomized Clinical Trial. JAMA Oncol 2021;7:1617-25. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Solomon BJ, Besse B, Bauer TM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol 2018;19:1654-67. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 2011;19:679-90. [Crossref] [PubMed]
- Zou HY, Friboulet L, Kodack DP, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell 2015;28:70-81. [Crossref] [PubMed]
- Johnson TW, Richardson PF, Bailey S, et al. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J Med Chem 2014;57:4720-44. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:1370-9. [Crossref] [PubMed]
- Shaw AT, Bauer TM, de Marinis F, et al. First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. N Engl J Med 2020;383:2018-29. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Gu FF, Zhang Y, Liu YY, et al. Lung adenocarcinoma harboring concomitant SPTBN1-ALK fusion, c-Met overexpression, and HER-2 amplification with inherent resistance to crizotinib, chemotherapy, and radiotherapy. J Hematol Oncol 2016;9:66. [Crossref] [PubMed]
- Lovly CM, McDonald NT, Chen H, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 2014;20:1027-34. [Crossref] [PubMed]
- Hrustanovic G, Olivas V, Pazarentzos E, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 2015;21:1038-47. [Crossref] [PubMed]
- Fan PD, Narzisi G, Jayaprakash AD, et al. YES1 amplification is a mechanism of acquired resistance to EGFR inhibitors identified by transposon mutagenesis and clinical genomics. Proc Natl Acad Sci U S A 2018;115:E6030-8. [Crossref] [PubMed]
- Dagogo-Jack I, Yoda S, Lennerz JK, et al. MET Alterations Are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer. Clin Cancer Res 2020;26:2535-45. [Crossref] [PubMed]
- Daniel C, Callens C, Melaabi S, et al. Acquired Exon 14 MET Mutation Associated With Resistance to Alectinib in a Patient With ALK-Rearranged NSCLC. JTO Clin Res Rep 2020;1:100082. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Minari R, Gnetti L, Lagrasta CA, et al. Emergence of a HER2-amplified clone during disease progression in an ALK-rearranged NSCLC patient treated with ALK-inhibitors: a case report. Transl Lung Cancer Res 2020;9:787-92. [Crossref] [PubMed]
- Sui A, Song H, Li Y, et al. BRAF V600E mutation as a novel mechanism of acquired resistance to ALK inhibition in ALK-rearranged lung adenocarcinoma: A case report. Medicine (Baltimore) 2021;100:e24917. [Crossref] [PubMed]
- Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014;346:1480-6. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Quintanal-Villalonga Á, Chan JM, Yu HA, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol 2020;17:360-71. [Crossref] [PubMed]
- Yamagata A, Yokoyama T, Fukuda Y, et al. Alectinib re-challenge in small cell lung cancer transformation after chemotherapy failure in a patient with ALK-positive lung cancer: A case report. Respir Med Case Rep 2021;33:101440. [Crossref] [PubMed]
- Xie Z, Gu Y, Lin X, et al. Unexpected favorable outcome to etoposide and cisplatin in a small cell lung cancer transformed patient: a case report. Cancer Biol Ther 2019;20:1172-5. [Crossref] [PubMed]
- Ball M, Christopoulos P, Kirchner M, et al. Histological and molecular plasticity of ALK-positive non-small-cell lung cancer under targeted therapy: a case report. Cold Spring Harb Mol Case Stud 2022;8:a006156. [Crossref] [PubMed]
- Zhang Y, Qin Y, Xu H, et al. Case Report: A Case Report of a Histological Transformation of ALK-Rearranged Adenocarcinoma With High Expression of PD-L1 to Squamous Cell Carcinoma After Treatment With Alectinib. Pathol Oncol Res 2021;27:637745. [Crossref] [PubMed]
- Fares AF, Lok BH, Zhang T, et al. ALK-rearranged lung adenocarcinoma transformation into high-grade large cell neuroendocrine carcinoma: Clinical and molecular description of two cases. Lung Cancer 2020;146:350-4. [Crossref] [PubMed]
- Kaiho T, Nakajima T, Iwasawa S, et al. ALK Rearrangement Adenocarcinoma with Histological Transformation to Squamous Cell Carcinoma Resistant to Alectinib and Ceritinib. Onco Targets Ther 2020;13:1557-60. [Crossref] [PubMed]
- Gong J, Gregg JP, Ma W, et al. Squamous Cell Transformation of Primary Lung Adenocarcinoma in a Patient With EML4-ALK Fusion Variant 5 Refractory to ALK Inhibitors. J Natl Compr Canc Netw 2019;17:297-301. [Crossref] [PubMed]
- Koyama K, Katsurada N, Jimbo N, et al. Overexpression of CD 133 and BCL-2 in non-small cell lung cancer with neuroendocrine differentiation after transformation in ALK rearrangement-positive adenocarcinoma. Pathol Int 2019;69:294-9. [Crossref] [PubMed]
- Park S, Han J, Sun JM. Histologic transformation of ALK-rearranged adenocarcinoma to squamous cell carcinoma after treatment with ALK inhibitor. Lung Cancer 2019;127:66-8. [Crossref] [PubMed]
- Sim J, Kim H, Hyeon J, et al. Anaplastic lymphoma kinase (ALK)-expressing Lung Adenocarcinoma with Combined Neuroendocrine Component or Neuroendocrine Transformation: Implications for Neuroendocrine Transformation and Response to ALK-tyrosine Kinase Inhibitors. J Korean Med Sci 2018;33:e123. [Crossref] [PubMed]
- Ou SI, Lee TK, Young L, et al. Dual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. Pitfall of solely relying on liquid re-biopsy? Lung Cancer 2017;106:110-4. [Crossref] [PubMed]
- Levacq D, D'Haene N, de Wind R, et al. Histological transformation of ALK rearranged adenocarcinoma into small cell lung cancer: A new mechanism of resistance to ALK inhibitors. Lung Cancer 2016;102:38-41. [Crossref] [PubMed]
- Fujita S, Masago K, Katakami N, et al. Transformation to SCLC after Treatment with the ALK Inhibitor Alectinib. J Thorac Oncol 2016;11:e67-72. [Crossref] [PubMed]
- Caumont C, Veillon R, Gros A, et al. Neuroendocrine phenotype as an acquired resistance mechanism in ALK-rearranged lung adenocarcinoma. Lung Cancer 2016;92:15-8. [Crossref] [PubMed]
- Cha YJ, Cho BC, Kim HR, et al. A Case of ALK-Rearranged Adenocarcinoma with Small Cell Carcinoma-Like Transformation and Resistance to Crizotinib. J Thorac Oncol 2016;11:e55-8. [Crossref] [PubMed]
- Takegawa N, Hayashi H, Iizuka N, et al. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann Oncol 2016;27:953-5. [Crossref] [PubMed]
- Miyamoto S, Ikushima S, Ono R, et al. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn J Clin Oncol 2016;46:170-3. [PubMed]
- Ueda S, Shukuya T, Hayashi T, et al. Transformation from adenocarcinoma to squamous cell lung carcinoma with MET amplification after lorlatinib resistance: A case report. Thorac Cancer 2021;12:715-9. [Crossref] [PubMed]
- Fukuda K, Takeuchi S, Arai S, et al. Epithelial-to-Mesenchymal Transition Is a Mechanism of ALK Inhibitor Resistance in Lung Cancer Independent of ALK Mutation Status. Cancer Res 2019;79:1658-70. [Crossref] [PubMed]
- Xie X, Chen X, Luo N, et al. Transformation of invasive lung adenocarcinoma with ALK rearrangement into pulmonary sarcomatoid carcinoma. J Cancer Res Clin Oncol 2022;148:2165-8. [Crossref] [PubMed]
- Urbanska EM, Sørensen JB, Melchior LC, et al. Changing ALK-TKI-Resistance Mechanisms in Rebiopsies of ALK-Rearranged NSCLC: ALK- and BRAF-Mutations Followed by Epithelial-Mesenchymal Transition. Int J Mol Sci 2020;21:2847. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Papadimitrakopoulou VA, Wu YL, Han JY, et al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol 2018;29:VIII741. [Crossref]
- Hartmaier RJ, Markovets A, Cho BC, et al. Tumor genomics in patients (pts) with advanced epidermal growth factor receptor mutant (EGFRm) non-small cell lung cancer (NSCLC) whose disease has progressed on first-line (1L) osimertinib therapy in the Phase II ORCHARD study. Cancer Res. 2022;82:LB078. [Crossref]
- Mc Leer A, Foll M, Brevet M, et al. Detection of acquired TERT amplification in addition to predisposing p53 and Rb pathways alterations in EGFR-mutant lung adenocarcinomas transformed into small-cell lung cancers. Lung Cancer 2022;167:98-106. [Crossref] [PubMed]
- Wang S, Xie T, Hao X, et al. Comprehensive analysis of treatment modes and clinical outcomes of small cell lung cancer transformed from epidermal growth factor receptor mutant lung adenocarcinoma. Thorac Cancer 2021;12:2585-93. [Crossref] [PubMed]
- Jin CB, Yang L. Histological transformation of non-small cell lung cancer: Clinical analysis of nine cases. World J Clin Cases 2021;9:4617-26. [Crossref] [PubMed]
- Li R, Jiang L, Zhou X, et al. Pseudo-small cell transformation in EGFR-mutant adenocarcinoma. Lung Cancer 2021;153:120-5. [Crossref] [PubMed]
- Vendrell JA, Quantin X, Serre I, et al. Combination of tissue and liquid biopsy molecular profiling to detect transformation to small cell lung carcinoma during osimertinib treatment. Ther Adv Med Oncol 2020;12:1758835920974192. [Crossref] [PubMed]
- Gu Y, Zhu X, Cao B, et al. Transformation to small cell lung cancer and activation of KRAS during long-term erlotinib maintenance in a patient with non-small cell lung cancer: A case report. Oncol Lett 2019;17:5219-23. [Crossref] [PubMed]
- Yang H, Liu L, Zhou C, et al. The clinicopathologic of pulmonary adenocarcinoma transformation to small cell lung cancer. Medicine (Baltimore) 2019;98:e14893. [Crossref] [PubMed]
- Lin MW, Su KY, Su TJ, et al. Clinicopathological and genomic comparisons between different histologic components in combined small cell lung cancer and non-small cell lung cancer. Lung Cancer 2018;125:282-90. [Crossref] [PubMed]
- Ferrer L, Giaj Levra M, Brevet M, et al. A Brief Report of Transformation From NSCLC to SCLC: Molecular and Therapeutic Characteristics. J Thorac Oncol 2019;14:130-4. [Crossref] [PubMed]
- Tokaca N, Wotherspoon A, Nicholson AG, et al. Lack of response to nivolumab in a patient with EGFR-mutant non-small cell lung cancer adenocarcinoma sub-type transformed to small cell lung cancer. Lung Cancer 2017;111:65-8. [Crossref] [PubMed]
- Suda K, Murakami I, Yu H, et al. Heterogeneity in Immune Marker Expression after Acquisition of Resistance to EGFR Kinase Inhibitors: Analysis of a Case with Small Cell Lung Cancer Transformation. J Thorac Oncol 2017;12:1015-20. [Crossref] [PubMed]
- Ahn S, Hwang SH, Han J, et al. Transformation to Small Cell Lung Cancer of Pulmonary Adenocarcinoma: Clinicopathologic Analysis of Six Cases. J Pathol Transl Med 2016;50:258-63. [Crossref] [PubMed]
- Hwang KE, Jung JW, Oh SJ, et al. Transformation to small cell lung cancer as an acquired resistance mechanism in EGFR-mutant lung adenocarcinoma: a case report of complete response to etoposide and cisplatin. Tumori 2015;101:e96-8. [Crossref] [PubMed]
- Ahmed T, Vial MR, Ost D, et al. Non-small cell lung cancer transdifferentiation into small cell lung cancer: A case series. Lung Cancer 2018;122:220-3. [Crossref] [PubMed]
- Santoni-Rugiu E, Grauslund M, Melchior LC, et al. Heterogeneous resistance mechanisms in an EGFR exon 19-mutated non-small cell lung cancer patient treated with erlotinib: Persistent FGFR3-mutation, localized transformation to EGFR-mutated SCLC, and acquired T790M EGFR-mutation. Lung Cancer 2017;113:14-7. [Crossref] [PubMed]
- Abdallah N, Nagasaka M, Abdulfatah E, et al. Non-small cell to small cell lung cancer on PD-1 inhibitors: two cases on potential histologic transformation. Lung Cancer (Auckl) 2018;9:85-90. [Crossref] [PubMed]
- Imakita T, Fujita K, Kanai O, et al. Small cell lung cancer transformation during immunotherapy with nivolumab: A case report. Respir Med Case Rep 2017;21:52-5. [Crossref] [PubMed]
- Sehgal K, Varkaris A, Viray H, et al. Small cell transformation of non-small cell lung cancer on immune checkpoint inhibitors: uncommon or under-recognized? J Immunother Cancer 2020;8:e000697. [Crossref] [PubMed]
- Bar J, Ofek E, Barshack I, et al. Transformation to small cell lung cancer as a mechanism of resistance to immunotherapy in non-small cell lung cancer. Lung Cancer 2019;138:109-15. [Crossref] [PubMed]
- Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377. [Crossref] [PubMed]
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. [Crossref] [PubMed]
- Baine MK, Rekhtman N. Multiple faces of pulmonary large cell neuroendocrine carcinoma: update with a focus on practical approach to diagnosis. Transl Lung Cancer Res 2020;9:860-78. [Crossref] [PubMed]
- George J, Walter V, Peifer M, et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat Commun 2018;9:1048. [Crossref] [PubMed]
- Popat S, Wotherspoon A, Nutting CM, et al. Transformation to "high grade" neuroendocrine carcinoma as an acquired drug resistance mechanism in EGFR-mutant lung adenocarcinoma. Lung Cancer 2013;80:1-4. [Crossref] [PubMed]
- Baglivo S, Ludovini V, Sidoni A, et al. Large Cell Neuroendocrine Carcinoma Transformation and EGFR-T790M Mutation as Coexisting Mechanisms of Acquired Resistance to EGFR-TKIs in Lung Cancer. Mayo Clin Proc 2017;92:1304-11. [Crossref] [PubMed]
- Belluomini L, Caliò A, Giovannetti R, et al. Molecular predictors of EGFR-mutant NSCLC transformation into LCNEC after frontline osimertinib: digging under the surface. ESMO Open 2021;6:100028. [Crossref] [PubMed]
- Le X, Desai NV, Majid A, et al. De novo pulmonary small cell carcinomas and large cell neuroendocrine carcinomas harboring EGFR mutations: Lack of response to EGFR inhibitors. Lung Cancer 2015;88:70-3. [Crossref] [PubMed]
- Wang Y, Shen YH, Ma S, et al. A marked response to icotinib in a patient with large cell neuroendocrine carcinoma harboring an EGFR mutation: A case report. Oncol Lett 2015;10:1575-8. [Crossref] [PubMed]
- De Pas TM, Giovannini M, Manzotti M, et al. Large-cell neuroendocrine carcinoma of the lung harboring EGFR mutation and responding to gefitinib. J Clin Oncol 2011;29:e819-22. [Crossref] [PubMed]
- Yanagisawa S, Morikawa N, Kimura Y, et al. Large-cell neuroendocrine carcinoma with epidermal growth factor receptor mutation: possible transformation of lung adenocarcinoma. Respirology 2012;17:1275-7. [Crossref] [PubMed]
- Harbour JW, Lai SL, Whang-Peng J, et al. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 1988;241:353-7. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Febres-Aldana CA, Chang JC, Ptashkin R, et al. Rb Tumor Suppressor in Small Cell Lung Cancer: Combined Genomic and IHC Analysis with a Description of a Distinct Rb-Proficient Subset. Clin Cancer Res 2022;28:4702-13. [Crossref] [PubMed]
- Ishioka K, Yasuda H, Hamamoto J, et al. Upregulation of FGF9 in Lung Adenocarcinoma Transdifferentiation to Small Cell Lung Cancer. Cancer Res 2021;81:3916-29. [Crossref] [PubMed]
- Xie T, Li Y, Ying J, et al. Whole exome sequencing (WES) analysis of transformed small cell lung cancer (SCLC) from lung adenocarcinoma (LUAD). Transl Lung Cancer Res 2020;9:2428-39. [Crossref] [PubMed]
- Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol 2012;25:S18-30. [Crossref] [PubMed]
- Quintanal-Villalonga A, Taniguchi H, Zhan YA, et al. Multiomic Analysis of Lung Tumors Defines Pathways Activated in Neuroendocrine Transformation. Cancer Discov 2021;11:3028-47. [Crossref] [PubMed]
- Borromeo MD, Savage TK, Kollipara RK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 2016;16:1259-72. [Crossref] [PubMed]
- Paz-Ares L, Chen Y, Reinmuth N, et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open 2022;7:100408. [Crossref] [PubMed]
- Liu SV, Reck M, Mansfield AS, et al. Updated Overall Survival and PD-L1 Subgroup Analysis of Patients With Extensive-Stage Small-Cell Lung Cancer Treated With Atezolizumab, Carboplatin, and Etoposide (IMpower133). J Clin Oncol 2021;39:619-30. [Crossref] [PubMed]
- Socinski MA, Smit EF, Lorigan P, et al. Phase III study of pemetrexed plus carboplatin compared with etoposide plus carboplatin in chemotherapy-naive patients with extensive-stage small-cell lung cancer. J Clin Oncol 2009;27:4787-92. [Crossref] [PubMed]
- Farago AF, Keane FK. Current standards for clinical management of small cell lung cancer. Transl Lung Cancer Res 2018;7:69-79. [Crossref] [PubMed]
- Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. [Crossref] [PubMed]
- Mahadevan NR, Knelson EH, Wolff JO, et al. Intrinsic Immunogenicity of Small Cell Lung Carcinoma Revealed by Its Cellular Plasticity. Cancer Discov 2021;11:1952-69. [Crossref] [PubMed]
- Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346-360.e7. [Crossref] [PubMed]
- Tatsumori T, Tsuta K, Masai K, et al. p40 is the best marker for diagnosing pulmonary squamous cell carcinoma: comparison with p63, cytokeratin 5/6, desmocollin-3, and sox2. Appl Immunohistochem Mol Morphol 2014;22:377-82. [Crossref] [PubMed]
- Kim MJ, Shin HC, Shin KC, et al. Best immunohistochemical panel in distinguishing adenocarcinoma from squamous cell carcinoma of lung: tissue microarray assay in resected lung cancer specimens. Ann Diagn Pathol 2013;17:85-90. [Crossref] [PubMed]
- Levin PA, Mayer M, Hoskin S, et al. Histologic Transformation from Adenocarcinoma to Squamous Cell Carcinoma as a Mechanism of Resistance to EGFR Inhibition. J Thorac Oncol 2015;10:e86-8. [Crossref] [PubMed]
- Kuiper JL, Ronden MI, Becker A, et al. Transformation to a squamous cell carcinoma phenotype of an EGFR-mutated NSCLC patient after treatment with an EGFR-tyrosine kinase inhibitor. J Clin Pathol 2015;68:320-1. [Crossref] [PubMed]
- Paik PK, Varghese AM, Sima CS, et al. Response to erlotinib in patients with EGFR mutant advanced non-small cell lung cancers with a squamous or squamous-like component. Mol Cancer Ther 2012;11:2535-40. [Crossref] [PubMed]
- Cheung AH, Tong JH, Chung LY, et al. EGFR mutation exists in squamous cell lung carcinoma. Pathology 2020;52:323-8. [Crossref] [PubMed]
- Zhao W, Choi YL, Song JY, et al. ALK, ROS1 and RET rearrangements in lung squamous cell carcinoma are very rare. Lung Cancer 2016;94:22-7. [Crossref] [PubMed]
- Krause A, Roma L, Lorber T, et al. Deciphering the clonal relationship between glandular and squamous components in adenosquamous carcinoma of the lung using whole exome sequencing. Lung Cancer 2020;150:132-8. [Crossref] [PubMed]
- Tata PR, Chow RD, Saladi SV, et al. Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Dev Cell 2018;44:679-693.e5. [Crossref] [PubMed]
- Mollaoglu G, Jones A, Wait SJ, et al. The Lineage-Defining Transcription Factors SOX2 and NKX2-1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment. Immunity 2018;49:764-779.e9. [Crossref] [PubMed]
- Quintanal-Villalonga A, Taniguchi H, Zhan YA, et al. Comprehensive molecular characterization of lung tumors implicates AKT and MYC signaling in adenocarcinoma to squamous cell transdifferentiation. J Hematol Oncol 2021;14:170. [Crossref] [PubMed]
- Meador CB, Cobb R, Hung Y, et al. EGFR-Mutant NSCLC with De Novo or Acquired Squamous Histology: Molecular Features and Clinical Outcomes. J Thorac Oncol 2021;16:S932-3. [Crossref]
- Filosso PL, Ruffini E, Asioli S, et al. Adenosquamous lung carcinomas: a histologic subtype with poor prognosis. Lung Cancer 2011;74:25-9. [Crossref] [PubMed]
- Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20:69-84. [Crossref] [PubMed]
- Hao Y, Bjerke GA, Pietrzak K, et al. TGFβ signaling limits lineage plasticity in prostate cancer. PLoS Genet 2018;14:e1007409. [Crossref] [PubMed]
- Aiello NM, Maddipati R, Norgard RJ, et al. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev Cell 2018;45:681-695.e4. [Crossref] [PubMed]
- Zhang W, Girard L, Zhang YA, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 2018;7:32-49. [Crossref] [PubMed]
- Piper-Vallillo AJ, Sequist LV, Piotrowska Z. Emerging Treatment Paradigms for EGFR-Mutant Lung Cancers Progressing on Osimertinib: A Review. J Clin Oncol 2020; Epub ahead of print. [Crossref] [PubMed]


