
High expression of transcription factor POU2F1 confers improved survival on smokers with lung adenocarcinoma: a retrospective study of two cohorts
Highlight box
Key findings
• High expression of POU2F1 improves outcome of smokers with adenocarcinoma of the lung.
What is known and what is new?
• POU2F1 is frequently deregulated in cancer and its role in other malignant entities such as breast cancer, gastric cancer and prostate cancer has been described.
• The role of POU2F1 in non-small cell lung cancer was now analyzed and its high expression resulted in improved survival of adenocarcinoma smokers, that otherwise frequently present with loss-of-function mutations.
What is the implication, and what should change now?
• To evaluate for clinical relevance, mechanisms of (inter-) action of POU2F1 in NSCLC should be analyzed, especially focusing on pharmacological interaction and cell survival.
Introduction
Lung cancer remains the leading cause of cancer-related mortality, and non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer patients (1,2). Several studies have reported a negative impact of cigarette-smoking on prognosis of lung cancer patients (3,4). Overall tumor mutational burden is manifold higher in NSCLC tumors of smokers compared to never-smokers (3,5-8), and recently, the genomic landscape of NSCLC in smokers has been unveiled as so called “tobacco signature” (5,8,9). In NSCLC of non-smokers, epidermal growth factor receptor (EGFR) mutations and tumor protein 53 (TP53) mutations are most common (8) and in adenocarcinoma (ADC) of the lung targetable genetic changes are frequently found including EGFR mutations (i.e., 71.6% in non-smokers vs. 40.4% in smokers) and anaplastic lymphoma kinase (ALK) rearrangements (i.e., 7.0% in non-smokers vs. 3.4% in smokers) (3). Moreover, non-smoking squamous cell carcinoma (SCC) patients may harbor an EGFR mutation in up to 8% of the cases, but in smokers its prevalence is at only 2.5%. While tobacco-induced genetic damages as predictor for targetable driver mutations has already been applied to treatment guidelines for NSCLC (10), the identification of novel biomarkers for smokers with NSCLC remains challenging. Solely, activating mutations of Kirsten rat sarcoma virus (KRAS) are more prominent in smokers with ADC than in non-smokers (i.e., 14.0% vs. 3.4%, respectively) (3) and can be targeted by sotorasib that has been approved for p.G12C-mutated KRAS-driven pre-treated NSCLC (11). However, the clinical efficacy of KRAS-inhibitors falls short of expectations (12). Other than KRAS, most of the smoking-associated mutations comprise loss-of-function of tumor-suppressive deoxyribonucleic acid (DNA)-damage repair genes, such as excision repair complementary complex 5 and 8 (ERCC5, ERCC8), DNA ligase 4 (LIG4), mutY DNA glycosylase (MUTYH), DNA polymerase beta (POLB), DNA repair protein REV1 as well as G/T mismatch-specific thymine DNA glycosylase (TDG) (9). While some of these genetic aberrations might predict responsiveness to platinum-based chemotherapies (13-16), none of them have been translated into targeted NSCLC therapies yet. Thus, discovery of novel targets for the treatment of smoking-related lung cancer is of primary importance.
The Pit-1, Oct1/2, Unc-86 (POU) domain class 2 transcription factor 1 (POU2F1) is a bipotential transcriptional regulator of induced and repressed states (9). POU2F1 is widely expressed in healthy as well as in malignant tissues (17) and a versatile acting transcription factor in cancer (18,19). Whereas in other malignant entities (20-22), including breast (23,24), gastric (25), cervical (26), and prostate cancer (27), an oncogenic role of POU2F1 has been described, its role in patients with NSCLC remains to be determined.
Here, we evaluated the prognostic role of POU2F1 in NSCLC, retrospectively analyzing protein and gene expression data of two large patient cohorts as well as its functional impact on human A549 lung cancer cell line. We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-714/rc).
Methods
Study collectives
To generate a hypothesis on POU2F1’s prognostic role in NSCLC, we retrospectively analyzed 311 patients, diagnosed, and surgically treated for non-small cell lung cancer (NSCLC) between December 1998 and November 2004 at the Thoracic Surgery department of the St. Georg’s Clinic Ostercappeln. Study approval by the Ethics Committees of Osnabrück, Münster and Mainz were obtained for the collection of paraffin-embedded tissue samples for biomarker testing. Individuals gave informed consent before participating in this anonymized retrospective analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Patients under 18 years of age, with an unknown smoking status, with stage IV disease, with an incomplete resection status (R1- or R2-resection) or with an indeterminate histological growth pattern (i.e., NSCLC not otherwise specified) were excluded from the primary analysis. Due to possible interaction with POU2F1 expression, exclusion of 94 neoadjuvant treated cases resulted in a final cohort size of 217 patients (see Figure 1A). Gathered clinical data included age at NSCLC diagnosis, gender, Eastern Cooperative Oncology Group (ECOG) performance status at diagnosis, smoking status and—if attainable—packyears, neoadjuvant treatment regimen, adjuvant treatment regimen, date of surgery, histopathological diagnosis, overall tumor stage, pT-, pN- and pM-status, tumor grading, resection status (R0, R1, or R2), date and status of relapse, as well as date and status of death or last contact.
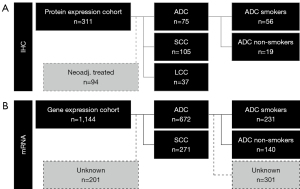
With regard to histopathology, 75 of 217 patients presented adenocarcinoma (ADC) growth pattern and 105 of 217 had squamous cell carcinoma (SCC) histology. The remaining 37 patients were diagnosed with large cell carcinoma (LCC). Staging was performed according to the sixth edition of Tumor Nodule Metastasis (TNM) system proposed by the “Union Internationale Contre le Cancer” (UICC) (28). An update towards later TNM staging system editions was not possible due to ethical concerns regarding patient privacy and data protection. Moreover, complete tissue specimen were paraffinized and no fresh frozen tissues were available. Hence, mutations in the EGFR- (29), and BRAF- (30), as well as translocations and fusions in ALK- (31), and ROS1- (32) gene loci could retrospectively not be assessed.
We used the online messenger ribonucleic acid meta-analysis database ‘kmplot.com’ for NSCLC as a validation cohort to evaluate the prognostic effect of POU2F1 gene expression (affymetrix Probe ID 227254_at) on the mRNA level in 672 ADC patients and 271 SCC patients (see Figure 1B). Regarding prior history of smoking, subcohort analyses of the ADC cohort were performed for 140 non-smokers and 231 smokers (33).
Protein expression analysis
The surgically resected primary tumor tissues were analyzed via 4 µm-thick formalin-fixed paraffin-embedded (FFPE) tissue microarrays (TMA). Each patient was represented by three punch cores from the original tumor specimen (34). Immunohistochemistry (IHC) was performed via the peroxidase-conjugated avidin-biotin method, as described previously (35). In brief, we used the rabbit polyclonal IgG anti-POU2F1 antibody ab15112 (Abcam, Catalog Number: ab15112) as primary antibody and immunoreactions were detected via the biotinylated secondary antibody (Roche/Ventana OptiView DAB IHC Detection Kit, Catalog Number: 760-700). Positive controls were performed on human tonsil tissues, negative controls on human healthy lung tissue. All IHC results were examined independently by three observers (ABS, GE, and BH) who were blinded to the patients’ clinical information. A Leica DM4B and an Olympus BX51 microscope with an average magnification of ×200 were used. The labeling indices were determined including all carcinoma cells within the tissue cores. For each punch core, the percentage of tumoral POU2F1 stain (0–100%) and the staining intensity from 0–3, following Remmele et al. (36), were assessed. Analyses were performed using the immunoreactive score (IRS) (37) deriving semiquantitative immunohistochemistry analysis. For the IRS, an ordinal category of the percentage of nuclear positive tumor cells (i.e., 0: no positive cells, 1: <10% positive cells, 2: 10–50% positive cells, 3: 51–80% positive cells, 4: >80% positive cells) is multiplied by the staining intensity (0, 1, 2, 3) and categorized as negative (0–1), mildly positive (2–3), moderately positive (4–8) and strongly positive (9–12). For statistical analysis, negative and mildly positive specimens (IRS ≤3) were summarized as ‘POU2F1 low’ and moderate to strongly positive ones (IRS 4–12) were summarized as ‘POU2F1 high’. Samples with discordant assessment results were re-evaluated until a consensus was reached.
Gene expression analysis
mRNA-based survival analyses were performed using the plotter on ‘kmplot.com’. Based on lung cancer microarray mRNA analysis (38), data from the “Cancer Biomedical Informatics Grid” (caBIG, ‘cabig.cancer.gov’), “Gene Expression Omnibus” (GEO, ‘ncbi.nlm.nig.gov/geo’) and “The Cancer Genome Atlas” (TCGA, ‘cancergenome.nih.gov’) were merged to allow for meta-survival analysis on mRNA expression in NSCLC patients (33), available at URL https://kmplot.com/analysis/ (date last accessed: 10/09/2022). To select for probes regarding POU2F1 mRNA, we chose the affymetrix Probe ID 227254_at, resulting in the above-mentioned cohort sizes (Figure 1B). The Kaplan-Meier plotter tool divides the cohort into patient groups with high versus low POU2F1 expression by median mRNA gene expression of the respective gene set (39).
Ectopic expression and CRISPR-Cas9 system-mediated knockdown of POU2F1 in human NSCLC cells
We designed two independent single guide RNAs (sgRNAs) targeting POU2F1 (V2 POU2F1 sgRNA1-2), using the website http://crispr.mit.edu/. sgRNAs were cloned into the lentiCRISPR V2 vector (Addgene, Watertown, MA 02472, USA). Lentiviral transduction of the human NSCLC cell line A549 was carried out as described previously (40). Ectopic expression of POU2F1 in A549 cells was performed via retroviral transduction as described previously (41). pBABE-POU2F1 was a kind gift from Dean Tantin. Ectopic expression and knockdown of POU2F1 (compared to controls) were confirmed via western blot.
Colony formation and proliferation assays
Colony formation and proliferation assays were performed as described previously (40). In brief, human NSCLC A549 cells were plated in methylcellulose (MethoCult H4034 Optimum, STEMCELL Technologies Germany, 50933 Cologne, Germany). Colonies were scored after 5–7 days. To assess proliferation, we used the CTG-assay (CellTiter-Glo® Luminescent Cell Viability Assay, Promega Germany, 69190 Walldorf, Germany) according to the manufacturer’s instructions. All experiments were performed independently at least three times.
Statistical analysis
To describe the cohort and cell culture analyses, we used mean, standard deviation (SD), median, interquartile range (IQR; Q1–Q3), 95% confidence interval (CI) as well as raw count and frequencies. Two-fold associations between categorical variables were analyzed via Fisher’s exact test or Chi-square test, if applicable. Continuous and ordinal variables were tested using either unpaired t-test or Mann-Whitney-U test, depending on the normality of the data. If applicable, two-tailed P values were recorded.
Overall survival (OS) included the time [days (protein expression cohort) or months (gene expression cohort)] between histopathological diagnosis and death or censoring. Progression-free survival (PFS) was defined as time [days (protein expression cohort) or months (gene expression cohort)] between histopathological diagnosis and first relapse, progress after initial treatment, death, or censoring (last visit alive, lost to follow-up), depending on the first chronological appearance. Univariate survival analyses compared OS and PFS between groups by using Log rank tests. Kaplan-Meier plots helped to visualize survival differences. We performed univariate Cox Regression analyses using the inclusion variable selection. Hazard ratios (HR) are presented with 95% CI. Due to a small sample size, a multivariate survival analysis was not performed in the protein expression cohort. Due to patient privacy, ‘kmplot.com’ extracted data did likewise not allow for multivariate survival testing. Data collection as well as calculations were performed using IBM® SPSS® Statistics Version 27 (released 2020, IBM Corp., Armonk, NY, USA). The Kaplan-Meier plotter online tool (‘kmplot.com’) automatically analyzes data by using the statistical software R with its underlying ‘survplot’ command of the survival Bioconductor package (33,42). Data of the Kaplan-Meier plotter online tool (‘kmplot.com’) were transferred to IBM® SPSS® Statistics via text output of the underlying raw data. Survival analyses in IBM® SPSS® Statistics were thus performed, as mentioned before. The local significance level was set to 0.05. Due to the explorative character of the protein-expression analysis, an adjustment to multiplicity was not determined.
Results
Baseline characteristics of the protein expression analysis cohort
Baseline characteristics of the patient cohort analyzed for nuclear POU2F1 protein expression are shown in Table 1. Via histopathology, a total of 217 patients were subdivided into 48.8% SCC, 34.6% ADC, and 17.1% LCC subtype. Male gender (79.7%) was more frequent than female gender. Among the selected cases, 76.9% of the patients were smokers. All patients were primarily treated by surgery and no neoadjuvant treatment was performed. According to the 6th edition TNM staging protocol (28) for lung cancer, 150 patients were classified as stage I NSCLC, 43 patients as stage II, and 24 patients as stage III. Two thirds of the tissue specimens showed high-grade tumors (G3/4). Median progression free survival (PFS) in this cohort was 1,321 days, median overall survival (OS) was 1,592 days and median follow-up was 2,744 days.
Table 1
| Characteristics | Items | Entire cohort | POU2F1 low | POU2F1 high | P value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total n=217 | %* | n=136 | %* | n=81 | %* | |||||
| Age (years) | Mean (± SD) | 66.79 (±8.23) | 65.74 (±8.41) | 68.54 (±7.66) | 0.013a | |||||
| Median (Q1–Q3) | 67.26 (62.40–72.16) | |||||||||
| Sex | Male | 173 | 79.7 | 102 | 75.0 | 71 | 87.7 | 0.035b | ||
| Female | 44 | 20.3 | 34 | 25.0 | 10 | 12.3 | ||||
| ECOG | 0 | 26 | 12.0 | 18 | 13.2 | 8 | 9.9 | 0.956c | ||
| I | 175 | 80.6 | 106 | 77.9 | 69 | 80.6 | ||||
| II–III | 16 | 7.4 | 12 | 8.8 | 4 | 4.9 | ||||
| Smoking status (n*=216) | Non-smoker | 50 | 23.1 | 30 | 22.1 | 20 | 25.0 | 0.621b | ||
| Smoker | 166 | 76.9 | 106 | 77.9 | 60 | 75.0 | ||||
| Histopathology | Squamous cell (SCC) | 105 | 48.4 | 60 | 44.1 | 45 | 55.6 | 0.011d | ||
| Adeno (ADC) | 75 | 34.6 | 57 | 41.9 | 18 | 22.2 | ||||
| Large cell (LCC) | 37 | 17.1 | 19 | 14.0 | 18 | 22.2 | ||||
| Grade (n*=214) | G1 | 2 | 0.9 | 2 | 1.5 | 0 | 0.0 | 0.009c | ||
| G2 | 63 | 29.4 | 45 | 33.6 | 18 | 22.5 | ||||
| G3 | 119 | 55.6 | 74 | 55.2 | 45 | 56.3 | ||||
| G4 | 30 | 14.0 | 13 | 9.7 | 17 | 21.3 | ||||
| Resection | R0 | 212 | 97.7 | 135 | 99.3 | 77 | 95.1 | 0.046c | ||
| R1 | 4 | 1.8 | 1 | 0.7 | 3 | 3.7 | ||||
| R2 | 1 | 0.5 | 0 | 0.0 | 1 | 1.2 | ||||
| UICC6 pT | pT1 | 65 | 30.0 | 42 | 30.9 | 23 | 28.4 | 0.848c | ||
| pT2 | 129 | 59.4 | 77 | 56.6 | 52 | 64.2 | ||||
| pT3 | 15 | 6.9 | 11 | 8.1 | 4 | 4.9 | ||||
| pT4 | 8 | 3.7 | 6 | 4.4 | 2 | 2.5 | ||||
| UICC6 pN | pN0 | 164 | 75.6 | 101 | 74.3 | 63 | 77.8 | 0.542c | ||
| pN1 | 40 | 18.4 | 26 | 19.1 | 14 | 17.3 | ||||
| pN2 | 13 | 6.0 | 9 | 6.6 | 4 | 4.9 | ||||
| UICC6 cM | cM0 | 217 | 100.0 | 136 | 100.0 | 81 | 100.0 | |||
| UICC6 pTNM | Stage I | 150 | 69.1 | 91 | 66.9 | 59 | 72.8 | 0.245c | ||
| Stage II | 43 | 19.8 | 26 | 19.1 | 17 | 21.0 | ||||
| Stage III | 24 | 11.1 | 19 | 14.0 | 5 | 6.2 | ||||
| PFS (days) | Median (95% CI) | 1,321 (946.4–1,695.6) | 1,322 (980.9–1,663.1) | 1,117 (188.6–2,045.3) | 0.902e | |||||
| OS (days) | Median (95% CI) | 1,592 (1,119.7–2,064.3) | 1,460 (941.6–1,978.4) | 1,657 (677.8–2,636.2) | 0.958e | |||||
| Follow-up (days) | Median (95% CI) | 2,744 (2,586.2–2,901.8) | 2,808 (2,641.7–2,974.3) | 2,558 (2,349.8–2,766.2) | 0.065e | |||||
%*, % of non-missing values; n*, non-missing values; P values calculated by a, Student’s t-test; b, Fisher’s exact test; c, Mann-Whitney-U test; d, Chi-square test; e, log-rank test. ECOG, eastern cooperative oncology group performance status; pT, pathological tumor status; pN, pathological lymph node status; cM, clinical metastasis status; PFS, progression-free survival; OS, overall survival; UICC, Union Internationale Contre le Cancer.
Correlations of clinical and histological parameters with POU2F1 protein expression
Immunoreactive score (IRS) (36,37) differentiated between low POU2F1 (i.e., IRS 0–3 =62.7% of the cohort) and high POU2F1 expression (i.e., IRS 4–12 =37.3%) (see Figure 2 for staining intensity). High POU2F1 expression was significantly associated with male gender (P=0.035), higher age (68.54 vs. 65.74 years, P=0.013) and high grade (G3/4) NSCLC tumors (P=0.009) (see Table 1). Moreover, high POU2F1 protein expression was also found more frequently in LCC and less frequently in ADC (P=0.011). Of marginal statistical significance, we found high POU2F1 expression more often in R1/2 resected tumors (P=0.046). Nevertheless, POU2F1 expression was neither associated with higher pT (P=0.848), pN (P=0.542) nor with pTNM staging (P=0.245), nor with smoking (P=0.621) or ECOG performance status (P=0.956). Finally, PFS (P=0.902) as well as OS (P=0.958) did not differ between patients with low versus high POU2F1 protein expression in the overall cohort (see Table 1).
High expression of POU2F1 confers improved survival in smokers with NSCLC adenocarcinoma
Regarding the entire patient cohort, expression of POU2F1 did not have any prognostic impact on survival of SCC patients, neither on protein [median OS: low expression 1,390 days vs. high expression 1,121 days, P=0.889; HR 0.97 (95% CI: 0.60–1.55)] nor on mRNA level [median OS: low expression 68.0 months vs. high expression 45.0 months, P=0.484; HR 1.12 (95% CI: 0.82–1.53)] (see Figure 3A,3B). However, ADC patients with high POU2F1 mRNA-expression levels had a significantly improved OS [median OS: low expression 79.9 months vs. high expression 136.3 months, P<0.001; HR 0.58 (95% CI: 0.45–0.75)], but only revealed an insignificant trend towards improved survival in the protein expression analysis [median OS: low expression 2,096 days vs. high expression not reached, P=0.154; HR 0.53 (95% CI: 0.22–1.28)] (see Figure 3C,3D). ADC patients also showed significantly longer PFS in the mRNA-based analysis (median PFS: low expression, n=222, 20.1 months vs. high expression, n=221, 42.7 months, P<0.001) but only a non-significant trend towards improved PFS in the evaluations of IHC-driven protein expression (median PFS: low expression 1,526 days vs. high expression not reached, P=0.183; data not shown). The analysis of the SCC subcohort resulted in equivalent PFS rates for patients with low vs. high POU2F1 expression in both, the protein-based (median PFS: low expression 1,322 days vs. high expression 921 days, P=0.707) as well as the mRNA-based evaluations (median PFS: low expression, n=70, 12.1 months vs. high expression, n=71, 12.6 months, P=0.470; data not shown).
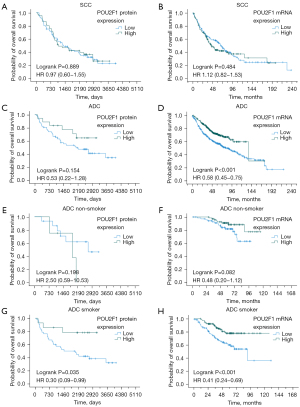
Next, we re-analyzed the ADC cohorts based on the smoking status of the patients. While for non-smokers, overexpression of POU2F1 was not associated with significant survival differences neither in the protein-based [median OS low 2,727 days vs. high 1,930 days, P=0.198; HR 2.50 (95% CI: 0.59–10.53)] nor in the mRNA-based analyses [median OS low not reached vs. high not reached, P=0.082; HR 0.48 (95% CI: 0.20–1.12)] (see Figure 3E,3F), but OS was significantly longer in ADC patients with a history of smoking in both cohorts [protein expression cohort: median OS low 1,460 days vs. high not reached, P=0.035; HR 0.30 (95% CI: 0.09–0.99); gene expression cohort: median OS low not reached vs. high not reached, P<0.001; HR 0.41 (95% CI: 0.24–0.69)] (see Figure 3G,3H). Thus, high expression of POU2F1 mediates improved outcome in smokers with ADC of the lung.
Association of PD-L1 protein expression and T-lymphocytic tumor infiltration with POU2F1 protein expression
PD-L1 is a marker of immune-evasion via the T-cellular PD-1 and tumoral PD-L1 receptor axis (43) and nowadays, inhibition of both receptors is common in systemic treatment of locally advanced (44) as well as metastasized NSCLC (45,46). Moreover, high tumor infiltration of CD8+ T-cytotoxic (Tc) cells is a marker of the patients’ immunoactivity and confers improved survival in PD-L1 low NSCLC tumor tissue (47). A positive correlation of POU2F1 expression with PD-L1 expression in NSCLC cell lines has been reported previously (48). Hence, we performed post-hoc correlation analyses of POU2F1 and PD-L1 protein expression (n=216) (43) as well as correlations with CD4+ T-helper (Th) cell, CD8+ Tc cell and FOXP3+ T-regulator (Treg) cell infiltration (n=205) (47) in our protein expression cohort.
However, regarding protein expression, we did not detect any association of tumoral PD-L1 positivity with high POU2F1 protein expression in the overall cohort (P=0.872). In the subgroup of ADC patients, raw counts indicated a slightly higher PD-L1 expression in POU2F1 overexpressing tumors, but statistical significance was not reached (P=0.246), potentially due to the small sample size. Smoking status did not have influence on tumoral PD-L1 expression intensity (both P=1.000 for smokers and non-smokers) (see Table S1). With respect to T-cellular infiltration, neither Th cell infiltration (all P>0.05, data not shown) nor Tc cell infiltration (all P>0.05, data not shown) could be correlated to nuclear POU2F1 protein expression. Yet, a significantly higher proportion of FOXP3+ Treg cells in POU2F1 overexpressing tumors was documented in the entire cohort (P=0.001) as well as in the smoker cohort (P=0.005), whereas in the ADC subcohort, no significant correlation between numbers of FOXP3+ Treg cells and POU2F1 expression could be detected (P=0.090) (see Table S2).
Thus, we did not determine a clear association between expression levels of POU2F1 and immune cell infiltration or immune escape of NSCLC tumors, respectively.
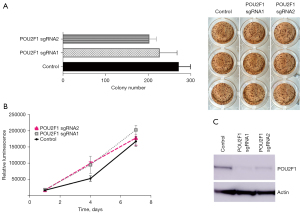
CRISPR-Cas9 system-mediated knockdown of POU2F1 does neither affect clonogenic growth nor proliferation of human NSCLC cells in vitro
To investigate the functional role of POU2F1 in lung cancer, we induced a CRISPR-Cas9 system-mediated knockdown of POU2F1 in human A549 NSCLC cells. Knockdown of POU2F1 neither had any significant impact on clonogenic growth of NSCLC cells (see Figure 4A) nor on proliferation after 1, 4 and 7 days of culture as assessed via CTG-assay and as compared to empty vector controls (see Figure 4B). POU2F1 protein knockdown was confirmed via western blot (see Figure 4C).
Overexpression of POU2F1 impairs clonogenic growth and proliferation of human NSCLC cells in vitro
Next, we induced overexpression of POU2F1 in human A549 NSCLC cells via retroviral transduction. Overexpression of POU2F1 resulted in both, significantly reduced clonogenic growth of NSCLC cells (see Figure 5A) as well as significantly impaired proliferation as assessed via CTG-assay after 1, 3 and 5 days of culture and as compared to empty vector-transduced control cells (see Figure 5B). Again, successful overexpression of POU2F1 was confirmed by western blot and compared to controls (see Figure 5C).

Thus, POU2F1 overexpression significantly impairs cell growth as well as proliferation of human NSCLC cells in vitro.
Discussion
Educational work on smoking-induced damage sustains primary prevention (49) and early detection via lung cancer screening in a prespecified high-risk cohort might help to reduce lung cancer-specific mortality (7,50). However, lung cancer is often detected in a locally advanced or even metastasized situation and hence, individual treatment options are needed to improve outcome (10). Smoking accounts for the majority of lung cancer cases, and patients with smoking-related lung cancer are known to harbor a higher tumor mutational burden (TMB) and a differential transcriptome (9), driving a malignant phenotype that differs from tumors of non-smoking patients (5,8). While in non-smokers, targetable driver mutations, such as ALK, EGFR, ROS1 and BRAF are more common (10), targetable genetic alterations in smokers are rare to date. Aberrant expression of transcription factors (TF) is frequent in NSCLC, and the differential tumor transcriptomes of smokers vs. non-smokers with lung cancer include a differential signature of tobacco exposure for various TF families (6,51).
POU2F1 is a member of the POU family of transcription factors that is widely expressed in human tissues and regulates cellular proliferation as well as immune modulation. Moreover, POU2F1 has been implicated in various types of epithelial cancers (18,20-27), while in patients with NSCLC its role had not been defined yet.
Here, we show that high expression of POU2F1 confers significantly improved outcome to smokers with ADC of the lung. Correspondingly, overexpression of POU2F1 significantly impaired both, clonogenic growth as well as proliferation of human A549 cells. Conversely, CRISPR-Cas9 system-mediated knockdown of POU2F1 did not affect proliferation or clonogenic growth of NSCLC cells.
The role of POU2F1 in cancer biology remains controversial since it has been identified as both, an oncogene as well as a tumor suppressor in different cancer entities (19,52-56). Results from cell culture and mouse model-based analyses suggested a POU2F1-induced immune-evasion via upregulation of tumoral PD-L1 expression in ADC of the lung (48). However, our study conducted in primary patient samples did not show any correlation between POU2F1 and PD-L1 expression, indicating that promotion of immune escape may not be the key function of POU2F1 in NSCLC patients. The variation between results is likely due to the usage of different experimental models since our analysis derived surgically obtained tissue whereas the previous work was performed using NSCLC cell lines.
At the transcriptional level, POU2F1 mediates both repression as well as activation of gene expression, even at the same target, by associating with different chromatin-modifying enzymes in a regulated manner (57). This may explain at least in part why aberrations of POU2F1 expression and/or actions can result in opposing ways in cancer. For instance, one of the known targets of POU2F1 is Gadd45a, a gene encoding a small protein belonging to the family of growth arrest- and DNA damage-inducible (GADD) proteins with versatile functions in control of cell cycle, apoptosis, and senescence (58-62). Depending on the oncogenic stress, Gadd45a can act as both, promoter or suppressor of cancer (63,64). Gadd45a is required for early embryonic cells to exit pluripotency and enter differentiation (65), and POU2F1-mediated transcription of GADD45 induces cellular growth arrest and apoptosis (66,67). Cigarette smoking induces oxidative stress, and POU2F1 is a known stress sensor modulating the activity of various genes orchestrating the cellular response to stress (68). Moreover, exposure to cigarette smoke downregulates the POU2F1 coactivator POU2AF1 (69,70).
Thus, we hypothesize that suppression of POU2F1 activity may promote progression of NSCLC and a more aggressive phenotype via repression of genes regulating growth arrest and apoptosis (e.g., Gadd45a) in a subset of smokers with NSCLC, whereas maintained expression of POU2F1 associates with improved survival of these patients. POU2F1 is post-translationally regulated by phosphorylation, ubiquitination and O-GlcNAcylation (71-74). While direct pharmacological targeting of POU2F1 with the intention to increase its expression and/or activity is out of reach, our data suggest POU2F1-regulated signaling pathways and genes as promising novel targets for future therapeutic approaches in NSCLC.
Conclusions
While POU2F1 protein and gene expression did not have impact on survival in patients with squamous cell growth pattern, especially POU2F1 high expressing tissue of smokers with adenocarcinoma of the lung was associated with improved outcome. In vitro A549 cell analyses stated the effect of reduced colony formation and proliferation upon POU2F1 overexpression but knock down of POU2F1 did not alter clonogenic growth or proliferation. While smokers often harbor loss-of-function mutations that are hardly targetable, POU2F1 expression might be a potential pharmaceutical target in this specific subset of NSCLC-patients. To fully understand its impact on NSCLC, further studies are needed to explore POU2F1’s cellular interactional behavior towards its co-players.
Acknowledgments
We thank Alessandro Marra, Department of Thoracic Surgery, Rems-Murr-Klinikum Winnenden, 71364 Winnenden, Germany as well as Ludger Hillejan, Department of Thoracic Surgery, Niels-Stensen-Kliniken, 48179 Ostercappeln, Germany for the primary collection of tissue specimens that were transferred onto TMAs. We thank Inka Buchroth, Lydia Fälker and Christin Fehmer from the Gerhard Domagk Institute of Pathology, University Hospital Muenster, for their excellent work on performing immunohistochemistry. Finally, we thank Dean Tantin from the Department of Pathology, Huntsman Cancer Institute of the University of Utah, Salt Lake City, UT 84112, USA, for providing the pBABE-POU2F1 plasmid.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-714/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-714/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-714/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-714/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was performed in concordance with the ethical approval obtained from the concerned Ethics Committees in Mainz, Osnabrück and Münster [Az 837.188.99 (2133), Az 4/71040/0/6 and Reg.Nr.: 4XMüller, respectively]. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Individuals gave informed consent before participating in this anonymized retrospective analysis.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: An overview. Int J Cancer 2021; Epub ahead of print. [Crossref] [PubMed]
- Schulze AB, Evers G, Kerkhoff A, et al. Future Options of Molecular-Targeted Therapy in Small Cell Lung Cancer. Cancers (Basel) 2019;11:690. [Crossref] [PubMed]
- Gou LY, Niu FY, Wu YL, et al. Differences in driver genes between smoking-related and non-smoking-related lung cancer in the Chinese population. Cancer 2015;121:3069-79. [Crossref] [PubMed]
- Sheikh M, Mukeriya A, Shangina O, et al. Postdiagnosis Smoking Cessation and Reduced Risk for Lung Cancer Progression and Mortality: A Prospective Cohort Study. Ann Intern Med 2021;174:1232-9. [Crossref] [PubMed]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Dai L, Jin B, Liu T, et al. The effect of smoking status on efficacy of immune checkpoint inhibitors in metastatic non-small cell lung cancer: A systematic review and meta-analysis. EClinicalMedicine 2021;38:100990. [Crossref] [PubMed]
- Caini S, Del Riccio M, Vettori V, et al. Quitting Smoking At or Around Diagnosis Improves the Overall Survival of Lung Cancer Patients: A Systematic Review and Meta-Analysis. J Thorac Oncol 2022;17:623-36. [Crossref] [PubMed]
- Boeckx B, Shahi RB, Smeets D, et al. The genomic landscape of nonsmall cell lung carcinoma in never smokers. Int J Cancer 2020;146:3207-18. [Crossref] [PubMed]
- Leong TL, Gayevskiy V, Steinfort DP, et al. Deep multi-region whole-genome sequencing reveals heterogeneity and gene-by-environment interactions in treatment-naive, metastatic lung cancer. Oncogene 2019;38:1661-75. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. [Crossref] [PubMed]
- Skoulidis F, Li BT, Dy GK, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 2021;384:2371-81. [Crossref] [PubMed]
- Cascetta P, Marinello A, Lazzari C, et al. KRAS in NSCLC: State of the Art and Future Perspectives. Cancers (Basel) 2022;14:5430. [Crossref] [PubMed]
- Jiang YH, Xu XL, Ruan HH, et al. The impact of functional LIG4 polymorphism on platinum-based chemotherapy response and survival in non-small cell lung cancer. Med Oncol 2014;31:959. [Crossref] [PubMed]
- Abdalkhalek ES, Wakeel LME, Nagy AA, et al. Variants of ERCC5 and the outcome of platinum-based regimens in non-small cell lung cancer: a prospective cohort study. Med Oncol 2022;39:152. [Crossref] [PubMed]
- Li M, Chen R, Ji B, et al. Role of ERCC5 polymorphisms in non small cell lung cancer risk and responsiveness/toxicity to cisplatin based chemotherapy in the Chinese population. Oncol Rep 2021;45:1295-305. [Crossref] [PubMed]
- Dong J, Wang X, Yu Y, et al. Association of Base Excision Repair Gene Polymorphisms with the Response to Chemotherapy in Advanced Non-Small Cell Lung Cancer. Chin Med J (Engl) 2018;131:1904-8. [Crossref] [PubMed]
- Jafek JL, Shakya A, Tai PY, et al. Transcription factor Oct1 protects against hematopoietic stress and promotes acute myeloid leukemia. Exp Hematol 2019;76:38-48.e2. [Crossref] [PubMed]
- Vázquez-Arreguín K, Tantin D. The Oct1 transcription factor and epithelial malignancies: Old protein learns new tricks. Biochim Biophys Acta 2016;1859:792-804. [Crossref] [PubMed]
- Vázquez-Arreguín K, Bensard C, Schell JC, et al. Oct1/Pou2f1 is selectively required for colon regeneration and regulates colon malignancy. PLoS Genet 2019;15:e1007687. [Crossref] [PubMed]
- Zhong Y, Huang H, Chen M, et al. POU2F1 over-expression correlates with poor prognoses and promotes cell growth and epithelial-to-mesenchymal transition in hepatocellular carcinoma. Oncotarget 2017;8:44082-95. [Crossref] [PubMed]
- Zhu HY, Cao GY, Wang SP, et al. POU2F1 promotes growth and metastasis of hepatocellular carcinoma through the FAT1 signaling pathway. Am J Cancer Res 2017;7:1665-79. [PubMed]
- Jin XM, Xu B, Zhang Y, et al. LncRNA SND1-IT1 accelerates the proliferation and migration of osteosarcoma via sponging miRNA-665 to upregulate POU2F1. Eur Rev Med Pharmacol Sci 2019;23:9772-80. [PubMed]
- Hwang-Verslues WW, Chang PH, Jeng YM, et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci U S A 2013;110:12331-6. [Crossref] [PubMed]
- Ma Y, Sekiya M, Kainoh K, et al. Transcriptional co-repressor CtBP2 orchestrates epithelial-mesenchymal transition through a novel transcriptional holocomplex with OCT1. Biochem Biophys Res Commun 2020;523:354-60. [Crossref] [PubMed]
- Liang Y, Zhang CD, Zhang C, et al. DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric Cancer 2020;23:212-27. [Crossref] [PubMed]
- Zhang R, Lu H, Lyu YY, et al. E6/E7-P53-POU2F1-CTHRC1 axis promotes cervical cancer metastasis and activates Wnt/PCP pathway. Sci Rep 2017;7:44744. [Crossref] [PubMed]
- Yamamoto S, Takayama KI, Obinata D, et al. Identification of new octamer transcription factor 1-target genes upregulated in castration-resistant prostate cancer. Cancer Sci 2019;110:3476-85. [Crossref] [PubMed]
- Tsim S, O'Dowd CA, Milroy R, et al. Staging of non-small cell lung cancer (NSCLC): a review. Respir Med 2010;104:1767-74. [Crossref] [PubMed]
- Chen YM. Update of epidermal growth factor receptor-tyrosine kinase inhibitors in non-small-cell lung cancer. J Chin Med Assoc 2013;76:249-57. [Crossref] [PubMed]
- Shea M, Costa DB, Rangachari D. Management of advanced non-small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches. Ther Adv Respir Dis 2016;10:113-29. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015;87:122-9. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241. [Crossref] [PubMed]
- Schmidt LH, Biesterfeld S, Kümmel A, et al. Tissue microarrays are reliable tools for the clinicopathological characterization of lung cancer tissue. Anticancer Res 2009;29:201-9. [PubMed]
- Schulze AB, Kuntze A, Schmidt LH, et al. High Expression of NT5DC2 Is a Negative Prognostic Marker in Pulmonary Adenocarcinoma. Cancers (Basel) 2022;14:1395. [Crossref] [PubMed]
- Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987;8:138-40. [PubMed]
- Fedchenko N, Reifenrath J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue - a review. Diagn Pathol 2014;9:221. [Crossref] [PubMed]
- Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Chen C, Wang X, Huang S, et al. Prognostic roles of Notch receptor mRNA expression in human ovarian cancer. Oncotarget 2017;8:32731-40. [Crossref] [PubMed]
- Mikesch JH, Schwammbach D, Hartmann W, et al. Reptin drives tumour progression and resistance to chemotherapy in nonsmall cell lung cancer. Eur Respir J 2018;52:1701637. [Crossref] [PubMed]
- Wenge DV, Felipe-Fumero E, Angenendt L, et al. MN1-Fli1 oncofusion transforms murine hematopoietic progenitor cells into acute megakaryoblastic leukemia cells. Oncogenesis 2015;4:e179. [Crossref] [PubMed]
- Schröder MS, Culhane AC, Quackenbush J, et al. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 2011;27:3206-8. [Crossref] [PubMed]
- Schmidt LH, Kümmel A, Görlich D, et al. PD-1 and PD-L1 Expression in NSCLC Indicate a Favorable Prognosis in Defined Subgroups. PLoS One 2015;10:e0136023. [Crossref] [PubMed]
- Faivre-Finn C, Vicente D, Kurata T, et al. Four-Year Survival With Durvalumab After Chemoradiotherapy in Stage III NSCLC—an Update From the PACIFIC Trial. J Thorac Oncol 2021;16:860-7. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Updated Analysis of KEYNOTE-024: Pembrolizumab Versus Platinum-Based Chemotherapy for Advanced Non–Small-Cell Lung Cancer With PD-L1 Tumor Proportion Score of 50% or Greater. J Clin Oncol 2019;37:537-46. [Crossref] [PubMed]
- Awad MM, Gadgeel SM, Borghaei H, et al. Long-Term Overall Survival From KEYNOTE-021 Cohort G: Pemetrexed and Carboplatin With or Without Pembrolizumab as First-Line Therapy for Advanced Nonsquamous NSCLC. J Thorac Oncol 2021;16:162-8. [Crossref] [PubMed]
- Schulze AB, Evers G, Görlich D, et al. Tumor infiltrating T cells influence prognosis in stage I-III non-small cell lung cancer. J Thorac Dis 2020;12:1824-42. [Crossref] [PubMed]
- Li F, Wang T, Huang Y. POU2F1 induces the immune escape in lung cancer by up-regulating PD-L1. Am J Transl Res 2021;13:672-83. [PubMed]
- Tanner NT, Kanodra NM, Gebregziabher M, et al. The Association between Smoking Abstinence and Mortality in the National Lung Screening Trial. Am J Respir Crit Care Med 2016;193:534-41. [Crossref] [PubMed]
- Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-iv21. [Crossref] [PubMed]
- Hammouz RY, Kostanek JK, Dudzisz A, et al. Differential expression of lung adenocarcinoma transcriptome with signature of tobacco exposure. J Appl Genet 2020;61:421-37. [Crossref] [PubMed]
- Boulon S, Dantonel JC, Binet V, et al. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol Cell Biol 2002;22:7769-79. [Crossref] [PubMed]
- Kakizawa T, Miyamoto T, Ichikawa K, et al. Silencing mediator for retinoid and thyroid hormone receptors interacts with octamer transcription factor-1 and acts as a transcriptional repressor. J Biol Chem 2001;276:9720-5. [Crossref] [PubMed]
- Magné S, Caron S, Charon M, et al. STAT5 and Oct-1 form a stable complex that modulates cyclin D1 expression. Mol Cell Biol 2003;23:8934-45. [Crossref] [PubMed]
- Préfontaine GG, Lemieux ME, Giffin W, et al. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol Cell Biol 1998;18:3416-30. [Crossref] [PubMed]
- Brockman JL, Schuler LA. Prolactin signals via Stat5 and Oct-1 to the proximal cyclin D1 promoter. Mol Cell Endocrinol 2005;239:45-53. [Crossref] [PubMed]
- Shakya A, Kang J, Chumley J, et al. Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J Biol Chem 2011;286:450-9. [Crossref] [PubMed]
- Hirose T, Sowa Y, Takahashi S, et al. p53-independent induction of Gadd45 by histone deacetylase inhibitor: coordinate regulation by transcription factors Oct-1 and NF-Y. Oncogene 2003;22:7762-73. [Crossref] [PubMed]
- Jin S, Fan F, Fan W, et al. Transcription factors Oct-1 and NF-YA regulate the p53-independent induction of the GADD45 following DNA damage. Oncogene 2001;20:2683-90. [Crossref] [PubMed]
- Takahashi S, Saito S, Ohtani N, et al. Involvement of the Oct-1 regulatory element of the gadd45 promoter in the p53-independent response to ultraviolet irradiation. Cancer Res 2001;61:1187-95. [PubMed]
- Dalvai M, Schubart K, Besson A, et al. Oct1 is required for mTOR-induced G1 cell cycle arrest via the control of p27(Kip1) expression. Cell Cycle 2010;9:3933-44. [Crossref] [PubMed]
- Mancini M, Leo E, Aluigi M, et al. Gadd45a transcriptional induction elicited by the Aurora kinase inhibitor MK-0457 in Bcr-Abl-expressing cells is driven by Oct-1 transcription factor. Leuk Res 2012;36:1028-34. [Crossref] [PubMed]
- Liebermann DA, Tront JS, Sha X, et al. Gadd45 stress sensors in malignancy and leukemia. Crit Rev Oncog 2011;16:129-40. [Crossref] [PubMed]
- Tront JS, Huang Y, Fornace AJ Jr, et al. Gadd45a functions as a promoter or suppressor of breast cancer dependent on the oncogenic stress. Cancer Res 2010;70:9671-81. [Crossref] [PubMed]
- Kaufmann LT, Niehrs C. Gadd45a and Gadd45g regulate neural development and exit from pluripotency in Xenopus. Mech Dev 2011;128:401-11. [Crossref] [PubMed]
- Bruemmer D, Yin F, Liu J, et al. Regulation of the growth arrest and DNA damage-inducible gene 45 (GADD45) by peroxisome proliferator-activated receptor gamma in vascular smooth muscle cells. Circ Res 2003;93:e38-47. [Crossref] [PubMed]
- Sheikh MS, Hollander MC, Fornance AJ Jr. Role of Gadd45 in apoptosis. Biochem Pharmacol 2000;59:43-5. [Crossref] [PubMed]
- Tantin D, Schild-Poulter C, Wang V, et al. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res 2005;65:10750-8. [Crossref] [PubMed]
- Gstaiger M, Knoepfel L, Georgiev O, et al. A B-cell coactivator of octamer-binding transcription factors. Nature 1995;373:360-2. [Crossref] [PubMed]
- Zhou H, Brekman A, Zuo WL, et al. POU2AF1 Functions in the Human Airway Epithelium To Regulate Expression of Host Defense Genes. J Immunol 2016;196:3159-67. [Crossref] [PubMed]
- Kang J, Gemberling M, Nakamura M, et al. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev 2009;23:208-22. [Crossref] [PubMed]
- Kang J, Goodman B, Zheng Y, et al. Dynamic regulation of Oct1 during mitosis by phosphorylation and ubiquitination. PLoS One 2011;6:e23872. [Crossref] [PubMed]
- Kang J, Shen Z, Lim JM, et al. Regulation of Oct1/Pou2f1 transcription activity by O-GlcNAcylation. FASEB J 2013;27:2807-17. [Crossref] [PubMed]
- Schild-Poulter C, Shih A, Tantin D, et al. DNA-PK phosphorylation sites on Oct-1 promote cell survival following DNA damage. Oncogene 2007;26:3980-8. [Crossref] [PubMed]


