Epigenetics in non-small cell lung cancer: from basics to therapeutics
Introduction
Lung cancer is a cancer of the modern man and only few cases date before the 20th century. By the mid-twentieth century it had swept the world, due to increased worldwide tobacco consumption (1). Even after widespread awareness about smoking in the last 4–5 decades, the lung cancer incidence has only plateaued and in the developed world, it still accounts for approximately 25% of cancer-related deaths. Detection of lung cancer at an early stage leads to a better prognosis; the 5-year survival for localized lung and bronchus cancer is 54.8%, compared to 27.4% for regional, and 4.2% for widely disseminated disease (1). Lung cancers are classified into small cell and non-small cell lung cancers (NSCLC) and approximately 80% are comprise of NSCLCs, which are classified into adenocarcinoma (AdC), adenosquamous carcinoma, squamous cell carcinoma (SqCC), and large cell carcinoma based on tumor histology, with 80% of NSCLC being either AdC or SqCC (2,3). Based on the molecular pathogenesis patterns and histologic classification, AdCs are the most common type to show common recurrent genomic gains and losses, and somatic mutations. These driver mutations have been extensively studied in AdCs and the most common ones mutated are the epidermal growth factor receptor (EGFR), KRAS, and anaplastic lymphoma kinase (ALK) oncogenes (2,3).
Over the past decade, epigenetics changes have been increasingly studied and used as markers for early cancer detection. Jean-Baptiste Lamarck first introduced the concept of epigenetics in his book Philosophie Zoologique more than 200 years ago, in which he called it “soft inheritance”. In 1939, Waddington defined epigenetics as “the causal interactions between genes and their products, which bring the phenotype into being” (4). Later Holiday defined epigenetics as heritable changes in gene expression that are not due to any DNA sequence alterations (5). Epigenetics consists of heritable modifications to the chromatin that influence gene expression and other DNA-dependent processes without directly altering the DNA coding sequence (5). These complex processes involve DNA methylation, microRNA regulation, and histone/nucleosome modifications. Mutations in epigenetic regulatory mechanisms and epigenetic pattern perturbations are implicated in many tumor types, including lung cancer, occurring through tumor suppressor gene silencing and oncogene activation. Epigenetic alterations are further linked to chemotherapy resistance (6,7). Unlike genetic mutations, epigenetic dysregulation is reversible and can be reversed by different pharmacologic approaches, the most common being hypomethylating agents and histone deacetylase inhibitors (HDACIs). Interestingly, while tumor heterogeneity represents a major challenge with targeted molecular therapies, broad re-modulation of the epigenome may address this problem through affecting multiple signaling pathways. The six FDA approved drugs that target the epigenome, their approval dates, and indications are summarized in Table 1.
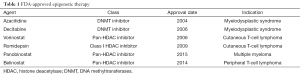
Full table
Epigenetic dysregulation in NSCLC
The initiation and progression of lung cancer is the result of permanent genetic alterations that include point mutations, deletions, translocations, amplifications, and epigenetic modifications that affect different aspects of chromatin-dependent processes, such as histone modifications, DNA methylation patterns, and microRNA regulation (8,9). DNA methylation plays a critical role in repressing gene expression and maintaining genomic stability by preventing recombination events between repetitive sequences (10). In eukaryotes, DNA methylation occurs in CpG dinucleotide islands which comprise roughly 1% of the human genome, but are present in over half of all human gene promoter sequences (11). In cancer cells, there is a dramatic reduction in cytosine methylation at these repetitive sequences, causing increased mitotic recombination and subsequent chromosomal instability (12-14). In addition, CpG islands of tumor suppression genes (TSGs) promoters are highly methylated leading to transcriptional repression, while other genes involved in processes such as DNA repair, apoptosis, the epithelial-mesenchymal transition (EMT), cellular movement and invasion, and metastasis are dysregulated by aberrant cytosine methylation (10,15). Further, hypomethylation of transposable element DNA can cause increased transposition within the genome, activating oncogenes and increasing chromosomal anomalies through insertional mutagenesis (16). Using the Illumina Infinium HumanMethylation27K array platform, Selamat et al. (17) interrogated the DNA methylation status of 27,578 CpG dinucleotides spanning 14,475 genes and identified more than 700 common differentially methylated genes. Additionally, accumulation of repressive histone markers is another hallmark of carcinogenesis, leading to chromatin compaction and gene expression repression. Histone deacetylases (HDACs) are often overexpressed in cancers and have become a major therapeutic target in recent years (18,19). HDAC overexpression can lead to TSG silencing and aberrant transcription due to altered expression/mutation of the genes encoding histone acetyltransferase (HAT) or HDAC enzymes or their binding partners, are clearly linked to carcinogenesis (20). This occurs in many human cancers, indicating that aberrant epigenetic acetylation activity is associated with cancer development (21-24).
DNA methylation
DNA methylation is the most studied epigenetic regulatory mechanism. CpG island methylation is completed by different DNA methyltransferases (DNMTs) that can lead to gene silencing. Three active DNMTs (DNMT1, DNMT3a, and DNMT3b) mediate the transfer of a methyl group from S-adenosyl-L-methionine to the CpG islands 5’-cytosine carbon (25-27). DNMT1 binds essentially to hemimethylated DNA and is primarily involved in the maintenance methylation after DNA replication. DNMT3a and b binds preferentially to unmethylated or hemimethylated DNA, and are responsible of de novo DNA methylation (28-30).
DNA methylation in lung cancer
DNMT overexpression is implicated in the pathogenesis of lung cancer. Elevated DNMT levels in lung cancer can result from transcriptional activator overexpression, loss of microRNAs that down-regulate the DNMTs, and/or impaired proteasomal DNMT degradation by hsp90 (21-23). Clinically, there is evidence that DNMT1 overexpression is associated with diminished survival in surgically resected lung cancer (31,32). Consistent with these findings, different TSGs are silenced by promoter hypermethylation in lung cancer (21). Many of these TSGs are involved in normal cellular function, such as cell cycle regulation (p16), DNA repair (MGMT), apoptosis (DAPK, caspase-8), regulation of Wnt signaling (APC), cell adhesion and invasion (E-cadherin, H-cadherin and tissue inhibitor of metalloproteinase-3), and suppression of invasion (CDH13, TIMP-3). For example, Brock et al. (22) observed that methylation of cdk2A, p16, CDH13, RASSF1A and APC correlated with recurrence following surgical resection of stage I NSCLC regardless of histology, stage, gender, or smoking history. Similarly, another study found that methylation of p16 and concomitant p16 expression loss coincides with reduced survival after early stage NSCLC resection (23). In parallel, IGFBP-3 methylation is linked to NSCLC cisplatin resistance (24).
Besides its role as a prognostic and predictive biomarker, DNA methylation has become a therapeutic target through DNMT enzyme inhibition. The two main DNMTs inhibitors that have been largely tested in the clinic are 5-azacitidine and decitabine (25). Following phosphorylation, 5-azacitidine is incorporated into DNA and RNA, followed by the covalent trapping of DNMTs to the DNA, leading to proteasomal degradation, and subsequent global DNA methylation reduction. The DNA damage and impaired DNA synthesis resulting from DNA-DNMT adducts is responsible for the direct cytotoxicity of these agents when used at higher doses. Unlike 5’-azacitidine, decitabine is not incorporated into the RNA and is specific only for DNA (26,27,33). The hypomethylating effects of these agents are best achieved at lower doses with a more prolonged administration (34). Pre-clinical models have shown antitumor activity for both agents through de-methylation and removal of repression on numerous TSGs, including p16 (35). Unfortunately, their use as single agent in clinical trials has showed limited success in lung cancer (36).
In a phase I/II trial, 15 patients with untreated advanced NSCLC were treated with high dose decitabine (200 to 660 mg/m2) administered as a continuous infusion over 8 hours. Although no objective response was seen, four patients experiences stable disease for more than 6 months, and three patients had a survival of at least 15 months, with one patient surviving 81 months. Due to hematopoietic toxicities, only one patient completed more than one cycle, which may have impacted the treatment efficacy (37,38). Another dose-escalation phase I trial conducted on 35 patients with solid tumor including 22 with lung cancer, investigated decitabine given at lower dose, administered over 72 hours continuous infusion. No objective response was seen, although three patients with squamous cell lung cancer had stable disease. Interestingly, pharmacodynamics studies revealed increased expression of p16, MAGE-3, and NY-ESO-1 in one-third of the patients (39). Further research will analyze the best sequence, dosage, treatment duration, and combination with other antineoplastic agents, as well as look for clinically relevant pharmacodynamic and predictive response biomarkers.
Smoking and DNA methylation
Some epigenetic alterations in lung cancer occur at greater frequency in smokers (i.e., p16, FHIT, RASSF1A mutations) and increase with increasing smoking duration/intensity (40-42). DNMT1 expression is elevated in smokers with lung cancer, likely due to tobacco-specific nitrosamines that reduce DNMT1 ubiquitination and degradation (21,43). Additionally smoking-induced chronic inflammation and increased reactive oxygen species generation, leading to increased DNA methylation (44). Damiani et al. (21) developed an in vitro model that mimics the field cancerization observed in chronic smokers and identified different epigenetic changes and their kinetics. Immortalized normal human bronchial epithelial cells (HBECs) were exposed for 12 weeks to two cigarette carcinogens; methylnitrosurea (MNU) and benzo(a)pyrene-diolepoxide 1 (BPDE). Stable knockdown of DNMT1, but not DNMT3 prevented cell transformation after exposure to these carcinogens. HBECs take a fibroblast-like mesenchymal appearance after 4 weeks of carcinogen exposure. Significant reductions in miR-200b and miR-200c, were observed at 4 weeks exposure and was sustained upon cell transformation at 12 weeks. Interestingly, these microRNAs are involved in regulating and inhibiting the EMT (35). Further studies revealed that expression of these EMT-regulating microRNAs are initially reduced by transcriptionally inactive chromatin at 4 weeks, followed later by cytosine methylation-mediated repression at their promoters (21,45).
On-going clinical trials
CC-486 is a novel oral azacitidine. An ongoing trial is examining the safety and efficacy of CC-486 in combination with pembrolizumab compared to pembrolizumab alone in previously treated advanced NSCLC (46). Another clinical trial is currently testing the combination of nab-paclitaxel with CC-486 when used in second line in advanced nonsquamous NSCLC (47). RRx-001 is a novel, broad-spectrum epigenetic anticancer agent that inhibits HDACs, DNMT1, and DNMT3a expression. Currently, it is being tested in lung cancer for its ability to sensitize the tumor to re-administration of a platinum doublet chemotherapy regimen (48). Another phase I trial is examining the tolerability and minimum effective dose of inhaled azacytidine (AZA) in NSCLC (49).
Histone modifications
Nucleosomes are chromosomal building blocks containing two molecules each of the core histones H2A, H2B, H3, and H4. DNA wraps around the nucleosomes octameric core approximately 1.8 times (50). The histone amino termini extend from the core, where they can be modified post-translationally by acetylation, phosphorylation, or methylation (51). Acetylation is regulated by opposing actions between HATs and HDACs (51). In humans there are 18 HDACs classified into four classes (I, II, III, IV) based on their homology to yeast HDAC’s, their subcellular localization, tissue specificity, and enzymatic activities (51).
HDAC inhibitors are emerging as novel anti-cancer agents due to their ability to kill cancer cells by inducing apoptosis, autophagy, cellular necrosis, ROS, cell cycle arrest, suppressing tumor angiogenesis, and exerting immunomodulatory effects. They activate both death-receptor and intrinsic mitochondrial pathways, lowering the overall tumor cell apoptotic threshold. They up-regulate pro-apoptotic genes involved in the death receptor pathway (i.e., TRAIL and DR5) and/or the intrinsic apoptotic pathway (Bax, Bak, and APAF1) and downregulate pro-survival genes (BCL-2 and XIAP). They also cause selective activation or induction of BH3-only proteins and hence initiate the intrinsic apoptotic pathway (52-54).
In addition to their direct anti-cancer effects, HDACIs strengthen the immune system by up-regulating the expression of MHC class I and II proteins, and co-stimulatory/adhesion molecules such as CD80, CD86, human leukocyte antigen (HLA)-DR, HLA-ABC, and intracellular adhesion molecule-1 (ICAM-1,28). HDACIs can also inhibit angiogenesis, a critical factor in tumor invasion and metastases (52,53). HDAC inhibitors may also enhance immune responses by altering the activities of immune cells, either directly or indirectly through cytokine secretion modulation (54).
The idea of HDAC inhibitors came through empirical screens, when molecular targets of agents that induce tumor differentiation were discovered, such as like butyrate, trichostatin A (TSA), and suberoylanilide hydroxamic acid (vorinostat) (52-54). By this theory, one can also deduce that HDAC in itself may be oncogenic, but there is no data demonstrating this. In contrast, knocking down of HDACs produces a range of antitumor effects. HDACI’s inhibit the growth of a wide variety of malignant cells in vitro, including lymphoma, myeloma, leukemia, and NSCLC, and inhibit the growth of a variety of solid tumors and hematological malignancies by both parenteral and oral administration, including prostate and breast cancers, leukemia, glioma, and lung cancer (52-54).
Histone modifications in lung cancer
Over the last decade, many studies have revealed epigenetic aberrations involving histone lung cancer modifications. Miyanaga et al. (54) tested 16 NSCLC cell lines with HDAC inhibitors including TSA and vorinostat, and both displayed antitumor activities in 50% of the NSCLC cell lines. They also conducted gene expression profiling and created a nine-gene classifier which predicts HDAC inhibitor drug sensitivities (54). Van Den Broeck et al. (46) showed that histone epigenetic modifications play a crucial role in lung carcinogenesis. Compared to normal lung cells, lung cancer cells displayed aberrant histone H4 modification patterns with hyperacetylation of H4K5/H4K8, hypoacetylation of H4K12/H4K16, and loss of H4K20 trimethylation. Their findings indicate an important role for histone H4 modifications and highlight H4K20me3 as a potential biomarker for the early detection of and therapeutic approaches to lung cancer (46). Seligson et al. (55) demonstrated that lower global levels of histone modifications are predictive of a more aggressive cancer phenotype in lung AdC. Additionally, the differential expression pattern of HATs and HDACs in the tumor samples, compared to the normal counterparts, can have potential therapeutic implications, such as eventual early tumor detection, prognostication, and the guiding of epigenetic-targeted therapies (56). HDAC1 gene expression appears to correlate with lung cancer progression, with strong HDAC1 and HDAC3 gene expression correlating with a poor prognosis in pulmonary AdC patients (57-59). HDAC3 was also seen elevated in 92% of tumor with SqCC histology using antibody microarrays for detection of target proteins (60). HDI-treated NSCLC cells down-regulated TNF-receptor-1 mRNA, protein levels, and surface protein expression, and consequently responded to TNF-treatment with attenuated NF-B nuclear translocation and DNA binding. HDIs, therefore, might beneficially contribute to tumor treatment, by reducing the responsiveness of tumor cells to the TNF-mediated activation of the NF-B pathway (61).
Treatment with TSA resulted in a dose-dependent reduction of H157 lung cancer cells by apoptosis with nuclear fragmentation and an increase in the sub-G0/G1 fraction. TSA initiated apoptosis by activation of the intrinsic mitochondrial and extrinsic/Fas/FasL system death pathways (62). TSA is also a powerful NSCLC cell radiosensitizer, enhancing G2/M cell cycle arrest, promoting apoptosis, and interfering with DNA damage repair, and synergistically triggering cell death when combined with other HDAC inhibitors, such as vorinostat (63,64). Vorinostat inhibits telomerase activity by reducing H-tert expression in A549 lung cancer cells (65). To explore the mechanisms by which vorinostat slows growth of lung cancer cells, changes of several key cell cycle and apoptosis proteins were examined. The cyclin-dependent kinase inhibitor p21 was upregulated in NCI-H520 and NCI-H460 cells treated with vorinostat. Prominent p21 induction was associated with G0-G1 cell-cycle arrest in vorinostat treated human lung cancer cells. p53 levels increased in NCI-H520 cells, partly explaining the increased p21, since p21 is a p53 transcriptional. C-myc levels decreased in both cell lines, indicating vorinostat exerts antiproliferative activity. Vorinostat also decreased bcl-2 expression in NCI-H460 cells (66).
Smoking and histone modifications
Cigarette smoke exposure to respiratory epithelia may also influence the histone modifications. Nickel cations present in tobacco smoke induce histone deacetylation and increase histone H3K9 dimethylation. Zhou et al. and others (67-69) found that smoke carcinogenic elements including nickel, chromate, and arsenite also induce H3K4 methylation.
Novel HDAC inhibitors
The novel cyclic amide-bearing hydroxamic acid-based HDAC inhibitors, SL142 and SL325, have shown greater HDAC inhibitory activity and lung cancer cell line viability-inhibition than vorinostat. These small molecules induce significant caspase-3 activity, indicating that they could induce lung cancer apoptosis (70). N-Hydroxy-4-(4-phenylbutyryl-amino) benzamide (HTPB), another novel HDAC inhibitor, caused significant lung cancer cell growth suppression by inducing cell cycle arrest, mitochondrial mediated apoptosis, disruption of F-actin dynamics, and inhibition of mitochondrial membrane potential (MMP)2 and MMP9. The effect was seen in vitro and in vivo (71). CG0006 a newly synthesized HDAC inhibitor, was assessed in a NCI-60 cancer cell panel and induced cell death by increased p21 and p27 expression (72). Other novel agents such as MGCD0103, OSU-HDAC 44, CI-944, MS-275, and LAQ824 were tested and show significant cytotoxic effects on lung cancer cells (73-80).
MicroRNA and microRNA silencing
MicroRNAs (miRs) are small, endogenous, single-stranded, noncoding RNAs of 20–22 nucleotides that regulate gene expression. More than 1,000 miRs have been identified and regulate more than one-third of coding mRNAs and each one can regulate hundreds of target mRNAs (81). Thus silencing of miRs by methylation can profoundly modulate tumor development and progression (82). Heller et al. (83) identified 33 miRs whose expression was increased in A549 cells (lung AdC) following demethylation treatment (83). miR-9-3, miR-34b and miR-126 are methylated in NSCLC and this is associated with an altered prognosis (83,84). miR-487b is commonly silenced by methylation in primary lung tumors and is reduced in respiratory epithelial cells and lung tumor-derived cell lines following tobacco smoke exposure (85). This finding reinforces past data which showing that smoking influences methylation and promotes lung cancer.
Combinatory epigenetic therapy
In normal cells, chromatin in the region of an actively transcribed tumor suppressor gene is typically in an open configuration, allowing transcription factor access. Carcinogenesis is associated with the epigenetic silencing of tumor suppressor genes, which may be secondary to DNA CpG island methylation and/or a closed chromatin configuration (6,86). Combined epigenetic therapy aims to reverse these alterations. Hypomethylating agents cause CpG island demethylation, allowing enhanced TSG transcription. On the other hand, HDAC inhibitors shift the chromatin to a more open configuration, favoring TSG transcription. While targeting each process alone has had disappointing effects on lung cancer, targeting both processes simultaneously may result in therapeutic synergism and enhanced TSG expression (87).
Combinatory hypomethylation and HDAC inhibition
Cameron et al. (88) demonstrated that while treatment with either TSA (an HDAC inhibitor) or decitabine alone has no effect on the TSG transcription, the drugs combined lead to synergistic reactivation of the TSG expression in colorectal carcinoma cells. Similarly, Boivin et al. (89) demonstrated that in lung cancer cell lines, AZA combined with the HDAC inhibitor phenylbutyrate exerted a greater DNA synthesis inhibition than either agent alone. Zhu et al. (90) 2001 found that lung cancer cell lines pre-treated with decitabine, show enhanced HDAC inhibitor-induced apoptosis and further enhanced histone acetylation. Similarly, AZA combined with the HDAC inhibitor entinostat inhibited lung cancer growth in an orthotopic mouse model, and caused the re-expression of p16, p21, and the pro-apoptotic gene PRC2. These results provide a sound scientific rationale for exploring combining hypomethylating agents with HDAC inhibitors in patients with advanced lung cancer (78,90).
Initial clinical trials using a combined approach failed to show significant response in patients with lung cancer:
- Decitabine and valproic acid, Chu et al. (91);
- 5-AZA and sodium phenylbutyrate, Lin et al. (92);
- Hydralazine and magnesium valproate, Candelaria et al. (93);
- Decitabine and vorinostat, Stathis et al. (94).
However, a more recent clinical trial combining AZA and entinostat in heavily pre-treated advanced NSCLC has revived interest in the combinatory approach. Strikingly, the achieved median overall survival was 6.4 months. Among the 34 evaluable patients, ten had stable disease for at least 12 weeks, one patient had complete response lasting 14 months and another patient had partial response lasting 8 months (87).
Role of epigenetic priming followed by conventional therapy
Interestingly, demethylation of four epigenetically silenced genes associated with lung cancer (APC, RASSF1a, CDH13, and CDKN2A) was detectable in serial plasma circulating DNA samples in these patients and was associated with improved progression free and overall survival (87). Another intriguing observation from this trial was a persistent clinical response after epigenetic therapy cessation and a notable clinical response to subsequent next anti-cancer treatments (cytotoxic chemotherapy and anti-PD1 monoclonal antibody) in many of these patients. Four out of 19 patients who received subsequent salvage therapy exhibited a major objective response; two patients survived 44 and 52 months respectively, after failing epigenetic treatment (95). This observation has raised an important hypothesis; epigenetic therapy may reprogram cancer cells and render them more susceptible to subsequent treatments. The above findings illustrate a new paradigm in cancer treatment—epigenetic priming. It consists of epigenetic modulator pretreatment prior to antineoplastic agent treatment. Currently many clinical trials are addressing this promising concept. An ongoing trial is currently randomizing patients with pretreated NSCLC to second line chemo alone vs. priming with 5-AZA and entinostat followed by second line chemotherapy. Another phase II trial is investigating the efficacy of nivolumab, a monoclonal antibody inhibitor of PD-1, after pre-treatment with 5-AZA and entinostat in patients with advanced NSCLC. This ongoing trial is based on recent findings that PD-L1 expression may be upregulated following treatment with AZA, increasing potentially the efficacy of anti-PD1 therapy (95). The combination of 5-AZA and entinostat is also being explored in the adjuvant setting with patients with resected stage I NSCLC, and comparing it to observation, the current standard of care after surgical resection in stage IA NSCLC (96).
Combinatory HDAC inhibition with other agents
Similarly, HDAC inhibitors show a more prominent effect when used in combination with other agents. Vorinostat has shown significant benefit when combined with carboplatin and paclitaxel in advanced lung cancer (96). Combinatorial TSA and etoposide treatment induced caspase-mediated AIF-dependent apoptotic cell death in non-small cell lung carcinoma cells. Genistein and carotene as nutritional factors in combination with TSA enhanced the cell growth arrest effect on A549 NSCLC cells (97). Combined treatment with low-dose vorinostat enhanced 5-FU drug-mediated cytotoxicity and resulted in synergistic effects, especially in 5-FU-resistant NSCLC cells. Vorinostat may overcome 5-FU resistance by down-regulating thymidylate synthase expression and up-regulating p21waf1/cip1 expression via histone acetylation at its promoter. This is the first report that vorinostat enhanced 5-FU sensitivity via the modulation of 5-FU metabolism in lung cancer cells and will facilitate future clinical investigations of combined chemotherapy and vorinostat in patients with NSCLC (66).
Millward et al. (98) combined vorinostat with a novel bi-cyclic proteasome inhibitor marizomib and tested advanced solid tumors, including NSCLC cells, and found highly synergistic antitumor activity. Although no responses were demonstrated using RECIST criteria, 61% of evaluable patients demonstrated stable disease with 39% having decreases in tumor measurements (98). Chien et al. (99) showed that vorinostat when combined with arsenic trioxide (ATO) acts synergistically to enhance in vitro and in vivo death of H1299 NSCLC cell. Seo et al. (100) combined suboptimal doses of Sulindac (NSAID) with vorinostat which resulted in growth suppression of A549 human NSCLC cells primarily via enhanced MMP collapse, release of cytochrome C, and caspase activation (100). Several on-going clinical trial using hypomethylating agents and HDAC inhibitors for the treatment of lung cancer are summarized Table 2.
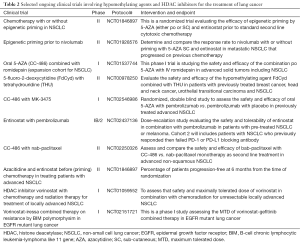
Full table
Novel therapeutic strategies: aerosol vidaza
After subcutaneous or intravenous administration, 5-AZA and decitabine are catabolized by cytidine deaminase in the liver, reducing the bioavailability of the drug to the lung. Aerosol delivery of these drugs may achieve higher concentrations in the pulmonary tissue by bypassing the hepatic first pass. Pharmacokinetic mice model had shown that aerosolized 5-AZA administration resulted in significant reduction of lung tumor burden and induction of global DNA demethylation at one-third of the comparable effective systemic dose (101).
Epigenetic biomarkers in lung cancer
Epigenetic changes as possible biomarkers for early lung cancer detection, diagnosis, prognostication, and the guiding of therapeutic options has recently been intensely studied, with heavy focus on DNA cytosine methylation, miR alterations, and histone modifications (6,8,9,46,50-52,54,81-85,102-104). Each of these epigenetic changes has specific testing methods and different degrees of clinical applicability. Presently most lung cancer epigenetic biomarkers are in development and will probably not have clinical application for several years (104).
DNA hypermethylation
DNA 5'-cytosine hypermethylation is an early lung carcinogenesis (6,8,9). Many genes are hypermethylated in lung cancer including p16, PAK3, NISCH, KIF1A, OGDHL, BRMS1, FHIT, CTSZ, CCNA1, NRCAM, LOX, MGMT, DOK1, SOX15, TCF21, DAPK, RAR, RASSF1, CYGB, MSX1, BNC1, CTSZ, and CDKN2A (105-118). The percent of hypermethylation for each gene varies, with some like p16 and MGMT hypermethylated in 100% of patients with pulmonary SqCC up the 3 years before cancer diagnosis (119). p16 inhibits cyclin-dependent kinases 4 and 6, which after binding cyclin D1, phosphorylate and inactivate the retinoblastoma tumor suppressor gene, blocking cell cycle progression (119). p16 is lost in ~70% of lung cancer cases, often by promoter methylation, promoting the G1 to S phase transition (119-121). Interestingly, p16 methylation occurs in normal-appearing epithelium from smokers and precursors lesions, and increases in frequency with the progression of the carcinogenic process (120). The specific mechanism(s) by which each gene hypermethylation event promotes cancer vary, but most involve repression of tumor suppressor genes with concomitant activation of genes promoting cell growth and cell cycle progression (105-122). Some of the genes hypermethylated in lung cancer and their functions are given in Table 3.

Full table
DNA hypermethylation in lung cancer patients can be detected in bronchoscopic washings/brushings, sputum samples, and blood (plasma and serum), all of which are less invasive and easier on the patient than a tumor biopsy. These techniques may also eventually be useful in the detection of very early lung tumors or newly recurring tumors not detectable by other methods (120). 5'-cytosine methylation is quantified predominantly by three different molecular methods:
- Methylation-sensitive restriction enzymes: most restriction endonucleases do not cut methylated DNA, while others only cut methylated DNA (137,138). There are many variations of this analysis, but they generally compare the activities of endonucleases that cut or will not cut methylated DNA, often using the isoschizomers MspI and HpaII which recognize CCGG with HpaII cutting blocked by either cytosine methylated and MspI activity blocked with the outer cytosine being methylated (137,138). Following endonuclease treatment, methylated or un-methylated DNA sequences are enriched and analyzed by PCR or DNA sequencing (120,137-139);
- Bisulfite conversion: under the correct conditions treatment of DNA with sodium bisulfite causes deamination of unmethylated cytosine to uracil, while leaving methylated cytosine intact. With PCR amplification the deaminated cytosine (a uracil) is copied into a thymine. The PCR products can be then analyzed by sequencing or mass spectrometry. Comparison of identical DNA samples with and without bisulfite allows analysis of the methylcytosine content and the specific methyl-cytosine moieties (140-142);
- Affinity purification methods: this technique uses either a methylcytosine-specific antibody or a tagged E. coli methyl-binding domain protein to immunoprecipitate methylated DNA (143-145). The resulting immunoprecipitates are commonly analyzed by next generation DNA sequencing (146).
A large number of studies have demonstrated that alteration in cytosine hypermethylation has diagnostic and prognostic value in lung cancer, and in some cases appears to predict treatment responses. A large number of studies have demonstrated that alteration in cytosine hypermethylation has diagnostic (82,102,105-122) and prognostic value in lung cancer, and in some cases appears to predict treatment responses (137-139,141-143,145-147). For example, Zhang et al. (147) examined the methylation of 20 TSGs in 78 NSCLCs compared to 50 matched plasma samples from individuals without cancer. A five-gene set (APC, RASSF1A, CHD13, KLK10, and DLEC1) showed significantly higher methylation in lung cancer patients and had a sensitivity of 83.64% and a specificity of 74.0% for cancer diagnosis in the Chinese population. The same study revealed that in a 64 lung cancer patient sample, patients with four or more concurrently methylated genes in a 15 gene panel (APC, CHD13, KLK13, DLEC1, RASSF1A, EFEMP1, SFRP1, RAR, p16INK4A, RUNX3, Hmlh1, DAPK, BRAC1, p14ARF) had a poorer progression-free two year survival of 13.8 months with four or more genes methylated compared to 17.8 months with less than four gene methylated. Last a study by Salazar et al. (148) revealed that lung cancer patients with an unmethylated plasma CHFR gene responded significantly better to EGFR tyrosine kinase inhibitors than those with a methylated CHFR gene, demonstrating that gene methylation might be useful in predicting therapy responses.
miR as biomarkers in lung cancer
miRs taken from sputum and blood may be useful lung cancer biomarkers (82-85,120). They are very stable in human plasma and have value in initial lung cancer detection (149-151). Bianchi et al. (150,151) developed a miR-34 group based test that detects lung cancer in 80% of asymptomatic high-risk individuals who are otherwise healthy smokers. Interestingly, this class of miRs has shown value in predicting lung cancer relapse, where low expression of these miRs were highly predictive of relapse (152). Similarly low miR-30a, miR-107, miR-138, miR-204, miR-32, miR-148b, miR-145, miR-224, miR-200c, miR-125b, and miR-375 predict a poor clinical outcome, and events such as increased lymph node metastasis and larger tumor size, while high miR-126, miR-21, miR-197, mi-150, and miR-141 levels predict also predict a poor outcome (153-167).
miRs have also been shown to have value in predicting therapeutic responses. For example, Zhao et al. (167) found that circulating miRs had value in predicting EGFR mutation, gefitinib sensitivity, and the patient’s prognosis (167). Last, miRs-33a and miR-124 may have prognostic value in lung cancer, with higher levels inhibiting the EMT transition and tumor metastasis, respectively (168,169). miRs are typically quantified by PCR amplification (149-169). Presently changes in miRs are seldom used clinically and developing reliable miR panels for clinical use in lung cancer diagnosis, prognostication, and treatment will take several more years (120).
Epigenetic changes in histones as lung cancer biomarkers
Histone modifications and changes in the expression patterns of HATs and HDACs may have value in early tumor detection, prognostication, and the guiding of epigenetic targeted therapies (55-61). Presently histone modifications and HATs and HDACs are used in the treatment of lung cancer, but their clinical use as lung cancer biomarkers and use in guiding therapy is several years away Presently histone modifications and HATs and HDACs are used in the treatment of lung cancer (59-74), but their clinical use as lung cancer biomarkers and use in guiding therapy is several years away (75-88,118).
Conclusion and future directions
Epigenetics plays an important role in early lung cancer development and progression. Recent studies have shown that methylation of TSGs correlates with the prognosis of resected early stage NSCLC, and this can be exploited to recognize which patient may benefit from adjuvant epigenetic therapy in order to reduce the risk of relapse after surgery. Since it can affect multiple pathways (21-24,28-32,46,54-61,67-69,82-85) that regulate all major properties of the cancer cell (105-122), targeting the epigenome may hold promise (137-148) in lung cancer therapy (153-169).
In spite of some disappointing clinical outcomes in earlier studies employing only epigenetic therapy, the field continues to evolve. Indeed, the remarkable responses to subsequent chemotherapy after epigenetic therapy with AZA and entinostat constitutes a paradigm shift in the management of metastatic NSCLC (76). There is a current trend to explore epigenetic priming agents to render lung cancer more susceptible to cytotoxic chemotherapy and immunotherapy. Finding predictive biomarkers to select patients who may derive benefits from epigenetic modulation and defining pharmacodynamic markers to gauge the efficacy of these agents, optimizing their effects, and their delivery sequence in conjunction with other antineoplastic agents, all constitute major challenges that need to be explored to move this promising field ahead.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- SEER. SEER Stat Fact Sheets: Lung and Bronchus Cancer 2015. Available online: http://seer.cancer.gov/statfacts/html/lungb.html
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. [Crossref] [PubMed]
- Shackelford RE, Vora M, Mayhall K, et al. ALK-rearrangements and testing methods in non-small cell lung cancer: a review. Genes Cancer 2014;5:1-14. [PubMed]
- Waddington CH. Preliminary Notes on the Development of the Wings in Normal and Mutant Strains of Drosophila. Proc Natl Acad Sci U S A 1939;25:299-307. [Crossref] [PubMed]
- Holliday R. The inheritance of epigenetic defects. Science 1987;238:163-70. [Crossref] [PubMed]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [PubMed]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92. [Crossref] [PubMed]
- Balgkouranidou I, Liloglou T, Lianidou ES. Lung cancer epigenetics: emerging biomarkers. Biomark Med 2013;7:49-58. [Crossref] [PubMed]
- Brzeziańska E, Dutkowska A, Antczak A. The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep 2013;40:309-25. [Crossref] [PubMed]
- Mehta A, Dobersch S, Romero-Olmedo AJ, et al. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev 2015;34:229-41. [Crossref] [PubMed]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010-22. [Crossref] [PubMed]
- Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A 2008;105:252-7. [Crossref] [PubMed]
- Kim M, Trinh BN, Long TI, et al. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res 2004;32:5742-9. [Crossref] [PubMed]
- Daskalos A, Nikolaidis G, Xinarianos G, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 2009;124:81-7. [Crossref] [PubMed]
- Zöchbauer-Müller S, Fong KM, Virmani AK, et al. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res 2001;61:249-55. [PubMed]
- Sargurupremraj M, Wjst M. Transposable elements and their potential role in complex lung disorder. Respir Res 2013;14:99. [Crossref] [PubMed]
- Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012;22:1197-211. [Crossref] [PubMed]
- Song J, Noh JH, Lee JH, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS 2005;113:264-8. [Crossref] [PubMed]
- Halkidou K, Gaughan L, Cook S, et al. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 2004;59:177-89. [Crossref] [PubMed]
- Mai A, Massa S, Rotili D, et al. Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev 2005;25:261-309. [Crossref] [PubMed]
- Damiani LA, Yingling CM, Leng S, et al. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res 2008;68:9005-14. [Crossref] [PubMed]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118-28. [Crossref] [PubMed]
- Sterlacci W, Tzankov A, Veits L, et al. A comprehensive analysis of p16 expression, gene status, and promoter hypermethylation in surgically resected non-small cell lung carcinomas. J Thorac Oncol 2011;6:1649-57. [Crossref] [PubMed]
- Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 2010;29:1681-90. [Crossref] [PubMed]
- Forde PM, Brahmer JR, Kelly RJ. New strategies in lung cancer: epigenetic therapy for non-small cell lung cancer. Clin Cancer Res 2014;20:2244-8. [Crossref] [PubMed]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002;21:5483-95. [Crossref] [PubMed]
- Patel K, Dickson J, Din S, et al. Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res 2010;38:4313-24. [Crossref] [PubMed]
- Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 2007;104:15805-10. [Crossref] [PubMed]
- Zhou Q, Agoston AT, Atadja P, et al. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res 2008;6:873-83. [Crossref] [PubMed]
- Lin RK, Wu CY, Chang JW, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res 2010;70:5807-17. [Crossref] [PubMed]
- Kim H, Kwon YM, Kim JS, et al. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer 2006;107:1042-9. [Crossref] [PubMed]
- Lin RK, Hsu HS, Chang JW, et al. Alteration of DNA methyltransferases contributes to 5'CpG methylation and poor prognosis in lung cancer. Lung Cancer 2007;55:205-13. [Crossref] [PubMed]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell 1980;20:85-93. [Crossref] [PubMed]
- Tsai HC, Li H, Van Neste L, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 2012;21:430-46. [Crossref] [PubMed]
- Merlo A, Herman JG, Mao L, et al. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995;1:686-92. [Crossref] [PubMed]
- Liu SV, Fabbri M, Gitlitz BJ, et al. Epigenetic therapy in lung cancer. Front Oncol 2013;3:135. [Crossref] [PubMed]
- Momparler RL, Ayoub J. Potential of 5-aza-2'-deoxycytidine #Decitabine# a potent inhibitor of DNA methylation for therapy of advanced non-small cell lung cancer. Lung Cancer 2001;34 Suppl 4:S111-5. [Crossref] [PubMed]
- Momparler RL, Bouffard DY, Momparler LF, et al. Pilot phase I-II study on 5-aza-2'-deoxycytidine (Decitabine) in patients with metastatic lung cancer. Anticancer Drugs 1997;8:358-68. [Crossref] [PubMed]
- Schrump DS, Fischette MR, Nguyen DM, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res 2006;12:5777-85. [Crossref] [PubMed]
- Toyooka S, Maruyama R, Toyooka KO, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 2003;103:153-60. [Crossref] [PubMed]
- Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 2001;61:3419-24. [PubMed]
- Liu Y, Lan Q, Siegfried JM, et al. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia 2006;8:46-51. [Crossref] [PubMed]
- Lin RK, Hsieh YS, Lin P, et al. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest 2010;120:521-32. [Crossref] [PubMed]
- O'Hagan HM, Wang W, Sen S, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 2011;20:606-19. [Crossref] [PubMed]
- Belinsky SA. Unmasking the lung cancer epigenome. Annu Rev Physiol 2015;77:453-74. [Crossref] [PubMed]
- Van Den Broeck A, Brambilla E, Moro-Sibilot D, et al. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 2008;14:7237-45. [Crossref] [PubMed]
- Safety and Efficacy Study of Nab®-Paclitaxel With CC-486 and Nab?-Paclitaxel Monotherapy as Second Line Treatment for Advanced Nonsquamous Non-small Cell Lung Cancer (abound2L). ClinicalTrials.gov 2014:NCT02250326. Available online: https://clinicaltrials.gov/ct2/show/NCT02250326?term=NCT02250326&rank=1
- RRx-001 in Small, Non-small Cell Lung Cancer, and Neuroendocrine Tumors Prior to Re-administration of Platinum Based Doublet Regimens (TRIPLE THREAT). ClinicalTrials.gov 2015:NCT02489903. Available online: https://clinicaltrials.gov/ct2/show/NCT02489903?term=NCT02489903&rank=1
- Inhaled Azacitidine in Patients With Advanced Non-small Cell Lung Cancer. ClinicalTrials.gov 2014:NCT02009436. Available online: https://clinicaltrials.gov/ct2/show/record/NCT02009436?term=NCT02009436&rank=1
- Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997;389:251-60. [Crossref] [PubMed]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997;389:349-52. [Crossref] [PubMed]
- Riggs MG, Whittaker RG, Neumann JR, et al. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 1977;268:462-4. [Crossref] [PubMed]
- Leder A, Leder P. Butyric acid, a potent inducer of erythroid differentiation in cultured erythroleukemic cells. Cell 1975;5:319-22. [Crossref] [PubMed]
- Miyanaga A, Gemma A, Noro R, et al. Antitumor activity of histone deacetylase inhibitors in non-small cell lung cancer cells: development of a molecular predictive model. Mol Cancer Ther 2008;7:1923-30. [Crossref] [PubMed]
- Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 2009;174:1619-28. [Crossref] [PubMed]
- Ozdağ H, Teschendorff AE, Ahmed AA, et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics 2006;7:90. [Crossref] [PubMed]
- Sasaki H, Moriyama S, Nakashima Y, et al. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer 2004;46:171-8. [Crossref] [PubMed]
- Minamiya Y, Ono T, Saito H, et al. Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer 2011;74:300-4. [Crossref] [PubMed]
- Minamiya Y, Ono T, Saito H, et al. Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumour Biol 2010;31:533-9. [Crossref] [PubMed]
- Bartling B, Hofmann HS, Boettger T, et al. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer 2005;49:145-54. [Crossref] [PubMed]
- Imre G, Gekeler V, Leja A, et al. Histone deacetylase inhibitors suppress the inducibility of nuclear factor-kappaB by tumor necrosis factor-alpha receptor-1 down-regulation. Cancer Res 2006;66:5409-18. [Crossref] [PubMed]
- Kim HR, Kim EJ, Yang SH, et al. Trichostatin A induces apoptosis in lung cancer cells via simultaneous activation of the death receptor-mediated and mitochondrial pathway? Exp Mol Med 2006;38:616-24. [Crossref] [PubMed]
- Zhang F, Zhang T, Teng ZH, et al. Sensitization to gamma-irradiation-induced cell cycle arrest and apoptosis by the histone deacetylase inhibitor trichostatin A in non-small cell lung cancer (NSCLC) cells. Cancer Biol Ther 2009;8:823-31. [Crossref] [PubMed]
- Seo SK, Jin HO, Woo SH, et al. Histone deacetylase inhibitors sensitize human non-small cell lung cancer cells to ionizing radiation through acetyl p53-mediated c-myc down-regulation. J Thorac Oncol 2011;6:1313-9. [Crossref] [PubMed]
- Li CT, Hsiao YM, Wu TC, et al. Vorinostat, SAHA, represses telomerase activity via epigenetic regulation of telomerase reverse transcriptase in non-small cell lung cancer cells. J Cell Biochem 2011;112:3044-53. [Crossref] [PubMed]
- Komatsu N, Kawamata N, Takeuchi S, et al. SAHA, a HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol Rep 2006;15:187-91. [PubMed]
- Stojanović D, Nikić D, Lazarević K. The level of nickel in smoker's blood and urine. Cent Eur J Public Health 2004;12:187-9. [PubMed]
- Zhou X, Li Q, Arita A, et al. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol 2009;236:78-84. [Crossref] [PubMed]
- Cantone L, Nordio F, Hou L, et al. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ Health Perspect 2011;119:964-9. [Crossref] [PubMed]
- Han S, Fukazawa T, Yamatsuji T, et al. Anti-tumor effect in human lung cancer by a combination treatment of novel histone deacetylase inhibitors: SL142 or SL325 and retinoic acids. PLoS One 2010;5:e13834. [Crossref] [PubMed]
- Shieh JM, Wei TT, Tang YA, et al. Mitochondrial apoptosis and FAK signaling disruption by a novel histone deacetylase inhibitor, HTPB, in antitumor and antimetastatic mouse models. PLoS One 2012;7:e30240. [Crossref] [PubMed]
- Hwang JJ, Kim YS, Kim MJ, et al. A novel histone deacetylase inhibitor, CG0006, induces cell death through both extrinsic and intrinsic apoptotic pathways. Anticancer Drugs 2009;20:815-21. [Crossref] [PubMed]
- Fournel M, Bonfils C, Hou Y, et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol Cancer Ther 2008;7:759-68. [Crossref] [PubMed]
- Gray J, Cubitt CL, Zhang S, et al. Combination of HDAC and topoisomerase inhibitors in small cell lung cancer. Cancer Biol Ther 2012;13:614-22. [Crossref] [PubMed]
- Tang YA, Wen WL, Chang JW, et al. A novel histone deacetylase inhibitor exhibits antitumor activity via apoptosis induction, F-actin disruption and gene acetylation in lung cancer. PLoS One 2010;5:e12417. [Crossref] [PubMed]
- Zhang Q, Feng W, Zhou H, et al. Advances in preclinical small molecules for the treatment of NSCLC. Expert Opin Ther Pat 2009;19:731-51. [Crossref] [PubMed]
- Niesen MI, Blanck G. Rescue of major histocompatibility-DR surface expression in retinoblastoma-defective, non-small cell lung carcinoma cells by the MS-275 histone deacetylase inhibitor. Biol Pharm Bull 2009;32:480-2. [Crossref] [PubMed]
- Belinsky SA, Grimes MJ, Picchi MA, et al. Combination therapy with vidaza and entinostat suppresses tumor growth and reprograms the epigenome in an orthotopic lung cancer model. Cancer Res 2011;71:454-62. [Crossref] [PubMed]
- Cuneo KC, Fu A, Osusky K, et al. Histone deacetylase inhibitor NVP-LAQ824 sensitizes human nonsmall cell lung cancer to the cytotoxic effects of ionizing radiation. Anticancer Drugs 2007;18:793-800. [Crossref] [PubMed]
- Beckers T, Burkhardt C, Wieland H, et al. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int J Cancer 2007;121:1138-48. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Ozsolak F, Poling LL, Wang Z, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev 2008;22:3172-83. [Crossref] [PubMed]
- Heller G, Weinzierl M, Noll C, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res 2012;18:1619-29. [Crossref] [PubMed]
- Watanabe K, Emoto N, Hamano E, et al. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer 2012;130:2580-90. [Crossref] [PubMed]
- Xi S, Xu H, Shan J, et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Invest 2013;123:1241-61. [Crossref] [PubMed]
- Esteller M. Cancer Epigenetics for the 21st Century: What's Next? Genes Cancer 2011;2:604-6. [Crossref] [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [Crossref] [PubMed]
- Cameron EE, Bachman KE, Myöhänen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999;21:103-7. [Crossref] [PubMed]
- Boivin AJ, Momparler LF, Hurtubise A, et al. Antineoplastic action of 5-aza-2'-deoxycytidine and phenylbutyrate on human lung carcinoma cells. Anticancer Drugs 2002;13:869-74. [Crossref] [PubMed]
- Zhu WG, Lakshmanan RR, Beal MD, et al. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res 2001;61:1327-33. [PubMed]
- Chu BF, Karpenko MJ, Liu Z, et al. Phase I study of 5-aza-2'-deoxycytidine in combination with valproic acid in non-small-cell lung cancer. Cancer Chemother Pharmacol 2013;71:115-21. [Crossref] [PubMed]
- Lin J, Gilbert J, Rudek MA, et al. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res 2009;15:6241-9. [Crossref] [PubMed]
- Candelaria M, Gallardo-Rincón D, Arce C, et al. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol 2007;18:1529-38. [Crossref] [PubMed]
- Stathis A, Hotte SJ, Chen EX, et al. Phase I study of decitabine in combination with vorinostat in patients with advanced solid tumors and non-Hodgkin's lymphomas. Clin Cancer Res 2011;17:1582-90. [Crossref] [PubMed]
- Wrangle J, Wang W, Koch A, et al. Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget 2013;4:2067-79. [Crossref] [PubMed]
- Azacitidine and Entinostat in Treating Patients With Stage I Non-Small Cell Lung Cancer That Has Been Removed By Surgery. ClinicalTrials.gov 2010:NCT01207726. Available online: https://clinicaltrials.gov/ct2/show/NCT01207726?term=NCT01207726&rank=1
- Shiau RJ, Chen KY, Wen YD, et al. Genistein and beta-carotene enhance the growth-inhibitory effect of trichostatin A in A549 cells. Eur J Nutr 2010;49:19-25. [Crossref] [PubMed]
- Millward M, Price T, Townsend A, et al. Phase 1 clinical trial of the novel proteasome inhibitor marizomib with the histone deacetylase inhibitor vorinostat in patients with melanoma, pancreatic and lung cancer based on in vitro assessments of the combination. Invest New Drugs 2012;30:2303-17. [Crossref] [PubMed]
- Chien CW, Yao JH, Chang SY, et al. Enhanced suppression of tumor growth by concomitant treatment of human lung cancer cells with suberoylanilide hydroxamic acid and arsenic trioxide. Toxicol Appl Pharmacol 2011;257:59-66. [Crossref] [PubMed]
- Seo SK, Jin HO, Lee HC, et al. Combined effects of sulindac and suberoylanilide hydroxamic acid on apoptosis induction in human lung cancer cells. Mol Pharmacol 2008;73:1005-12. [Crossref] [PubMed]
- Reed MD, Tellez CS, Grimes MJ, et al. Aerosolised 5-azacytidine suppresses tumour growth and reprogrammes the epigenome in an orthotopic lung cancer model. Br J Cancer 2013;109:1775-81. [Crossref] [PubMed]
- Li J, Li WX, Bai C, et al. Particulate matter-induced epigenetic changes and lung cancer. Clin Respir J 2015. [Epub ahead of print]. [PubMed]
- Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713-8. [Crossref] [PubMed]
- Liloglou T, Bediaga NG, Brown BR, et al. Epigenetic biomarkers in lung cancer. Cancer Lett 2014;342:200-12. [Crossref] [PubMed]
- Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med 2006;3:e486. [Crossref] [PubMed]
- Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet 2004;75:460-74. [Crossref] [PubMed]
- Smith LT, Lin M, Brena RM, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A 2006;103:982-7. [Crossref] [PubMed]
- Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307-12. [PubMed]
- Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007;3:1709-23. [Crossref] [PubMed]
- Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 2002;1:287-99. [Crossref] [PubMed]
- Toyota M, Ho C, Ohe-Toyota M, et al. Inactivation of CACNA1G, a T-type calcium channel gene, by aberrant methylation of its 5' CpG island in human tumors. Cancer Res 1999;59:4535-41. [PubMed]
- Estécio MR, Yan PS, Ibrahim AE, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res 2007;17:1529-36. [Crossref] [PubMed]
- Palmisano WA, Crume KP, Grimes MJ, et al. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res 2003;63:4620-5. [PubMed]
- Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res 2008;68:2661-70. [Crossref] [PubMed]
- Mungall AJ, Palmer SA, Sims SK, et al. The DNA sequence and analysis of human chromosome 6. Nature 2003;425:805-11. [Crossref] [PubMed]
- Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006;38:1378-85. [Crossref] [PubMed]
- Ito M, Ito G, Kondo M, et al. Frequent inactivation of RASSF1A, BLU, and SEMA3B on 3p21.3 by promoter hypermethylation and allele loss in non-small cell lung cancer. Cancer Lett 2005;225:131-9. [Crossref] [PubMed]
- Wang M, Vikis HG, Wang Y, et al. Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res 2007;67:93-9. [Crossref] [PubMed]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995;81:323-30. [Crossref] [PubMed]
- Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 1998;95:11891-6. [Crossref] [PubMed]
- Shackelford RE, Kaufmann WK, Paules RS. Cell cycle control, checkpoint mechanisms, and genotoxic stress. Environ Health Perspect 1999;107 Suppl 1:5-24. [Crossref] [PubMed]
- Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681-6. [Crossref] [PubMed]
- Brabender J, Usadel H, Metzger R, et al. Quantitative O(6)-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: associations with clinical outcome. Clin Cancer Res 2003;9:223-7. [PubMed]
- Gomes A, Reis-Silva M, Alarcão A, et al. Promoter hypermethylation of DNA repair genes MLH1 and MSH2 in adenocarcinomas and squamous cell carcinomas of the lung. Rev Port Pneumol 2014;20:20-30. [Crossref] [PubMed]
- Tang X, Khuri FR, Lee JJ, et al. Hypermethylation of the death-associated protein (DAP) kinase promoter and aggressiveness in stage I non-small-cell lung cancer. J Natl Cancer Inst 2000;92:1511-6. [Crossref] [PubMed]
- Kim DH, Nelson HH, Wiencke JK, et al. Promoter methylation of DAP-kinase: association with advanced stage in non-small cell lung cancer. Oncogene 2001;20:1765-70. [Crossref] [PubMed]
- Hopkins-Donaldson S, Ziegler A, Kurtz S, et al. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ 2003;10:356-64. [Crossref] [PubMed]
- Shivapurkar N, Toyooka S, Eby MT, et al. Differential inactivation of caspase-8 in lung cancers. Cancer Biol Ther 2002;1:65-9. [Crossref] [PubMed]
- Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res 2002;8:1178-84. [PubMed]
- Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer 2007;58:131-8. [Crossref] [PubMed]
- Kim DS, Kim MJ, Lee JY, et al. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer 2007;110:2785-92. [Crossref] [PubMed]
- Heller G, Fong KM, Girard L, et al. Expression and methylation pattern of TSLC1 cascade genes in lung carcinomas. Oncogene 2006;25:959-68. [Crossref] [PubMed]
- Grote HJ, Schmiemann V, Kiel S, et al. Aberrant methylation of the adenomatous polyposis coli promoter 1A in bronchial aspirates from patients with suspected lung cancer. Int J Cancer 2004;110:751-5. [Crossref] [PubMed]
- Virmani AK, Rathi A, Zöchbauer-Müller S, et al. Promoter methylation and silencing of the retinoic acid receptor-beta gene in lung carcinomas. J Natl Cancer Inst 2000;92:1303-7. [Crossref] [PubMed]
- Schmidt B, Liebenberg V, Dietrich D, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010;10:600. [Crossref] [PubMed]
- Sato K, Tomizawa Y, Iijima H, et al. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep 2006;15:129-35. [PubMed]
- Korch C, Hagblom P. In-vivo-modified gonococcal plasmid pJD1. A model system for analysis of restriction enzyme sensitivity to DNA modifications. Eur J Biochem 1986;161:519-24. [Crossref] [PubMed]
- Waalwijk C, Flavell RA. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res 1978;5:3231-6. [Crossref] [PubMed]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005;74:481-514. [Crossref] [PubMed]
- Tost J, Gut IG. Analysis of gene-specific DNA methylation patterns by pyrosequencing technology. Methods Mol Biol 2007;373:89-102. [PubMed]
- Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A 2005;102:15785-90. [Crossref] [PubMed]
- Ehrich M, Field JK, Liloglou T, et al. Cytosine methylation profiles as a molecular marker in non-small cell lung cancer. Cancer Res 2006;66:10911-8. [Crossref] [PubMed]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet 1994;6:236-44. [Crossref] [PubMed]
- Zhang X, Jacobsen SE. Genetic analyses of DNA methyltransferases in Arabidopsis thaliana. Cold Spring Harb Symp Quant Biol 2006;71:439-47. [Crossref] [PubMed]
- Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest 2005;85:1172-80. [Crossref] [PubMed]
- Jung M, Kadam S, Xiong W, et al. MIRA-seq for DNA methylation analysis of CpG islands. Epigenomics 2015;7:695-706. [Crossref] [PubMed]
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [Crossref] [PubMed]
- Salazar F, Molina MA, Sanchez-Ronco M, et al. First-line therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer 2011;72:84-91. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Bianchi F, Nicassio F, Veronesi G, et al. Circulating microRNAs: next-generation biomarkers for early lung cancer detection. Ecancermedicalscience 2012;6:246. [PubMed]
- Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503. [Crossref] [PubMed]
- Gallardo E, Navarro A, Viñolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009;30:1903-9. [Crossref] [PubMed]
- Tang R, Liang L, Luo D, et al. Downregulation of MiR-30a is Associated with Poor Prognosis in Lung Cancer. Med Sci Monit 2015;21:2514-20. [Crossref] [PubMed]
- Guo W, Zhang Y, Zhang Y, et al. Decreased expression of miR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int J Mol Med 2015;36:1720-6. [PubMed]
- Bai Y, Wang YL, Yao WJ, et al. Expression of miR-32 in human non-small cell lung cancer and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol 2015;8:824-9. [PubMed]
- Ge H, Li B, Hu WX, et al. MicroRNA-148b is down-regulated in non-small cell lung cancer and associated with poor survival. Int J Clin Exp Pathol 2015;8:800-5. [PubMed]
- Shen H, Shen J, Wang L, et al. Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed Pharmacother 2015;69:301-5. [Crossref] [PubMed]
- Zhu D, Chen H, Yang X, et al. Decreased microRNA-224 and its clinical significance in non-small cell lung cancer patients. Diagn Pathol 2014;9:198. [Crossref] [PubMed]
- Shao Y, Geng Y, Gu W, et al. Prognostic significance of microRNA-375 downregulation in solid tumors: a meta-analysis. Dis Markers 2014;2014:626185.
- Zhong KZ, Chen WW, Hu XY, et al. Clinicopathological and prognostic significance of microRNA-107 in human non small cell lung cancer. Int J Clin Exp Pathol 2014;7:4545-51. [PubMed]
- Han L, Zhang G, Zhang N, et al. Prognostic potential of microRNA-138 and its target mRNA PDK1 in sera for patients with non-small cell lung cancer. Med Oncol 2014;31:129. [Crossref] [PubMed]
- Kim MK, Jung SB, Kim JS, et al. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch 2014;465:463-71. [Crossref] [PubMed]
- Zhao W, Zhao JJ, Zhang L, et al. Serum miR-21 level: a potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med 2015;8:14759-63. [PubMed]
- Mavridis K, Gueugnon F, Petit-Courty A, et al. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer 2015;112:1527-35. [Crossref] [PubMed]
- Yin QW, Sun XF, Yang GT, et al. Increased expression of microRNA-150 is associated with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:842-6. [PubMed]
- Tejero R, Navarro A, Campayo M, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One 2014;9:e101899. [Crossref] [PubMed]
- Zhao Q, Cao J, Wu YC, et al. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am J Cancer Res 2015;5:1692-705. [PubMed]
- Yang L, Yang J, Li J, et al. MircoRNA-33a inhibits epithelial-to-mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer. Sci Rep 2015;5:13677. [Crossref] [PubMed]
- Zhang Y, Li H, Han J, et al. Down-regulation of microRNA-124 is correlated with tumor metastasis and poor prognosis in patients with lung cancer. Int J Clin Exp Pathol 2015;8:1967-72. [PubMed]


