
Case report: targeted sequencing improves the diagnosis of multiple synchronous lung cancers
Highlight box
Key findings
• Different lesions in the same case had different driver mutations, suggesting that the 2 lesions were driven by different molecular events. Therefore, targeted sequencing containing driver genes should be used for the diagnosis of multiple synchronous lung cancers.
What is known and what is new?
• The traditional diagnostic criteria for MPLC/IPM including the Martini and Melamed (MM) criteria and the comprehensive histologic assessment (CHA) criteria, mainly relies on histological comparison between multiple lesions.
• Targeted sequencing revealed the clonality status of these lesions and improved their diagnosis
What is the implication, and what should change now?
• Targeted sequencing containing driver genes should be used for the diagnosis of multiple synchronous lung cancers.
Introduction
Diagnosis of multiple pulmonary nodules has become more frequent due to the application of low dose computed tomography (LDCT) (1,2). It is crucial for clinicians to distinguish between satellite nodules, multiple primary lung cancer (MPLC), and intrapulmonary metastases (IPM), as differential diagnosis is important for both prognosis and treatment. The traditional diagnostic criteria for MPLC/IPM, including the Martini and Melamed (MM) criteria and the comprehensive histologic assessment (CHA) criteria (3,4), mainly relies on histological comparison between multiple lesions. However, these criteria remain controversial in clinical practice. It is difficult to distinguish accurately between MPLC and IPM when the histology of the lesions is similar (5).
Recently, 2 criteria were introduced by the American College of Chest Physicians (ACCP) and the International Association for the Study of Lung Cancer Staging (IASLC) (6,7). Both criteria emphasize the consideration of all available information, including the histologic features, immunohistochemistry (IHC), clinical features, and molecular features. Based on the ACCP criteria, multiple synchronous lung cancers with the same histology can be classified into satellite nodules (same lobe, no systemic metastases), MPLC [different lobes, no N2–N3 lymph node (LN) involvement or systemic metastases], and IPM (different lobes, N2–N3 LN involvement) (7). It is generally accepted that locally spreading satellite nodules arise from the corresponding primary tumor (8,9); however, the clonal origin of multiple synchronous lesions is still widely debated (10). In cases where the histologic features of the lesions are highly similar, the distinction between MPLC and IPM is facilitated by molecular test such as targeted sequencing. Next-generation sequencing (NGS) is the most commonly used targeted sequencing technology, which can simultaneously sequence millions of DNA fragments (or complementary DNA), enabling more sensitive, economical and high-throughput detection. As a result, NGS are receiving increasing attention as an aid to histopathological diagnosis, particularly in lung cancer, where molecular typing of lung cancer contributes to treatment selection. It is now integrated into routine clinical oncology practice, particularly in non-small cell lung cancer (11-16).
Here, we report 3 lung adenocarcinoma patients who presented with 2 lesions, with improved diagnosis based on targeted sequencing. We present the following article in accordance with the CARE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-155/rc).
Case presentation
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patients for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Patients and specimens
All enrolled patients and specimens were from the Pathology Department of Henan Cancer Hospital in China from May 2020 to December 2020. Clinicopathological information was also extracted from the medical records. All samples were formalin-fixed paraffin-embedded (FFPE) sections and reviewed by 2 independent experienced pathologists.
Targeted sequencing
Tumor DNA and paracancer DNA were extracted from FFPE specimens using the TIANamp FFPE DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. DNA libraries were constructed with the KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) and captured with a targeted panel of 1,238 genes (Tianjin Novogene Bioinformatics Technology Co., Ltd., Tianjin, China) according to the manufacturer’s protocol. Sequencing was performed using the HiSeq X-Ten platform (Illumina, San Diego, CA, USA).
After removing low-quality sequencing reads, clean reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Aligner (BWA, v0.7.8) in default mode (17). VarScan (v2.4.3; https://varscan.sourceforge.net/) and GATK (v4.1; https://gatk.broadinstitute.org/hc/en-us) were used to call somatic and germline single-nucleotide variants (SNV) and small insertions or deletions (InDel) mutations. For tumor somatic mutations, the minimum mutation allele frequencies (MAF) ≥1% were reported. The mutations were then annotated using SnpEff (18) + ANNOVAR (v4.3) (18,19) VEP + ANNOVAR (ensembl-vep 90.6). CNVkit (v0.9.9) and Delly (v0.8.7) were used to call copy number variations and gene fusion, respectively (20,21). Non-synonymous mutations annotated by ANNOVAR were utilized for phylogenetic tree construction using a standard approach as described previously (22). Mutations that were common to all lesions were deemed trunk mutations.
Case 1
A 61-year-old female [patient 1 (P1)] who presented with 2 pulmonary nodules including 1 in the right middle lobe (RML; P1A) and 1 in the right upper lobe (RUL; P1B) was admitted to our hospital in October 2020. She then received right bilobectomy(middle and lower lobe)and systematic LNs dissection. The 2 resected nodules were all diagnosed as invasive adenocarcinoma (IA) without vascular tumor thrombus, perineural invasion, and LN metastasis. Histopathology showed that the P1B was acinar subtype, whereas the P1A was acinar and papillary subtype (Figure 1A,1B). The 2 lesions were displayed different lobes and different histological subtypes and therefore tended to be classified as MPLC (Table 1). However, co-occurring mutations in EGFR (p.L858R) and TET2 (p. F352Lfs*16) were identified in the P1A and P1B lesions (Figure 1C), indicating that the 2 lesions were IPM. At the 14-month post-operative follow-up, the patient had no recurrence.
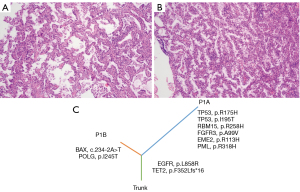
Table 1
| Case | Age (year) | Gender | Smoking | Sample | Location | Size (cm) | Histology | Subtype | Node staging | Histologic relationship | ACCP | Genomic prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 61 | Female | No | P1A | RML | 2.5×2×1.5 | IA | Acinar, papillary | N0 | Different | MPLC | IPM |
| P1B | RUL | 0.5 | IA | Acinar | ||||||||
| P2 | 61 | Female | No | P2A | RUL | 0.5×0.3×0.3 | MIA | Lepidic | N0 | Identical | Satellite nodules | MPLC |
| P2B | RUL | 0.7 | MIA | Lepidic | ||||||||
| P3 | 51 | Male | No | P3A | RUL | 1.4×1×1.2 | IA | Acinar, lepidic, papillary | N0 | Similar | Satellite nodules | MPLC |
| P3B | RUL | 1.2×0.7×0.7 | IA | Acinar, lepidic |
ACCP, American College of Chest Physicians; P, patient; RML, right middle lobe; IA, invasive adenocarcinoma; MPLC, multiple primary lung cancer; IPM, intrapulmonary metastases; RUL, right upper lobe; MIA, minimally invasive adenocarcinoma.
Case 2
A 61-year-old female (P2) with 2 pulmonary nodules (P2A and P2B) in the RUL was admitted to our hospital in December 2020. She then underwent RUL lobectomy resection and LNs dissection. Both resected nodules were diagnosed as minimally invasive adenocarcinoma (MIA) without lymphovascular invasion, perineural invasion, vascular tumor thrombus, and LN metastasis. Both nodules were of the lepidic subtype (Figure 2A,2B) and in the same lobe, therefore tended to be classified as satellite nodules (Table 1). However, the sequencing results showed that no mutation was shared in both lesions (Figure 2C). In addition, the oncogenic mutations BRAF p.G469A and MAP2K1 p.E102_I103del were detected in P2A and P2B, respectively. The MAP2K1 p.E102_I103del is mutually exclusive with other mutations that activate MAPK signaling and is therefore considered a driver mutation (23,24), whereas BRAF p.G469A is an oncogenic hotspot mutation in BRAF (25). Therefore, the 2 lesions were from independent clonal origins. The patient remained in good recovery without recurrence until the latest follow-up in January 2022 [disease-free survival (DFS) >13 months].

Case 3
A 51-year-old male (P3) underwent a health check-up at our hospital. Computed tomography (CT) showed 2 pulmonary nodules in the RUL. The patient underwent RUL lobectomy and systematic LNs dissection in May 2020. Histopathology showed 2 nodules of IA, without perineural invasion, vascular tumor thrombus, and LN metastasis. These 2 nodules were in the same lung lobe and had similar histologic subtypes (Figure 3A,3B) and were therefore considered satellite nodules (Table 1). However, molecular analysis showed that no shared somatic mutation was observed. Furthermore, the driver mutation of EGFR L858R was in 1 lesion, whereas the driver mutation of EGFR p.L747_T751del was detected in the other (Figure 3C). Given that these 2 mutations are usually exclusive in lung adenocarcinoma, we considered them MPLC. The patient has not relapsed (DFS >19 months) and remains stable.

Discussion
Multiple synchronous lung cancers account for 0.2–15% of lung cancers (26,27). However, the differentiation between MPLC and IPM/satellite nodule is a subject of debate. Clinically, MPLC need to be distinguished from IPM of lung cancer, and their treatment methods are quite different. A study (28) found that there was a significant difference in prognosis between MPLC and IM. Therefore, accurate identification of MPLC and IM is particularly important. Accumulating evidence suggests that analysis of somatic mutations has become an addition to improve the reliability of differentiation between MPLC and IPM (11-16,29,30). Previous studies mainly focused on the detection of hot spots and common driver genes to assist in the differential diagnosis of MPLA and IPM. NGS detection is helpful in differential diagnosis (31-34). In this study, we improved the diagnosis of MPLC or IPM in 3 patients by targeted sequencing. Of these, P1 was classified as MPLC according to the ACCP criteria, although molecular testing indicated that she was IPM. The other 2 patients (P2 and P3) were classified as satellite nodules according to the ACCP criteria; however, molecular testing showed that these 2 patients should be diagnosed with MPLC. In both cases, different lesions in the same case had different driver mutations (35), suggesting that the 2 lesions were driven by different molecular events. A limitation of this report is the short follow up period, and long-term outcomes of the patients require further follow up.
A previous report showed that up to 32% of all histologically confirmed MPLC were misclassified as IPM compared to molecular analysis (36). Here, additional molecular information provided by targeted sequencing allowed us to correctly identify these subtypes. Targeted sequencing can simultaneously detect multiple genes including driver genes. Compared to conventional diagnostic techniques of lung cancer, targeted gene sequencing can further clarify the molecular genetic map, assist in the identification of lung cancer subtypes, and distinguish the diagnosis of IPM and MPLC in patients with multiple nodules. At present, single gene assays, is mainly to detect EGFR, which is mainly used to guide treatment. While most lung adenocarcinomas are not EGFR mutated, the differential diagnosis between IPM and MPLC is limited and does not fully reflect the molecular characteristics of patient. So targeted sequencing that includes more comprehensive driver genes is more advantageous.
Conclusions
In the future, molecular genetic maps can be established by targeted sequencing to better assist patients with multiple nodules in the differential diagnosis of IPM and MPLC. This report can enrich knowledge about the differentiation between MPLC and IPM/satellite nodules. Targeted sequencing containing driver genes should be used for the diagnosis of multiple synchronous lung cancers and provide a more comprehensive basis for the diagnosis and treatment of patients.
Acknowledgments
The authors thank the patients for their permissions to publish their cases.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-155/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-155/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-155/coif). LM and SS are from Novogene Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patients for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- van der Aalst CM, Ten Haaf K, de Koning HJ. Lung cancer screening: latest developments and unanswered questions. Lancet Respir Med 2016;4:749-61. [Crossref] [PubMed]
- Yang D, Liu Y, Bai C, et al. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett 2020;468:82-7. [Crossref] [PubMed]
- Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70:606-12. [Crossref] [PubMed]
- Girard N, Deshpande C, Lau C, et al. Comprehensive histologic assessment helps to differentiate multiple lung primary nonsmall cell carcinomas from metastases. Am J Surg Pathol 2009;33:1752-64. [Crossref] [PubMed]
- Murphy SJ, Harris FR, Kosari F, et al. Using Genomics to Differentiate Multiple Primaries From Metastatic Lung Cancer. J Thorac Oncol 2019;14:1567-82. [Crossref] [PubMed]
- Detterbeck FC, Franklin WA, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposed Criteria to Distinguish Separate Primary Lung Cancers from Metastatic Foci in Patients with Two Lung Tumors in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:651-65.
- Kozower BD, Larner JM, Detterbeck FC, et al. Special treatment issues in non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e369S-99S.
- Gazdar AF, Minna JD. Multifocal lung cancers--clonality vs field cancerization and does it matter? J Natl Cancer Inst 2009;101:541-3. [Crossref] [PubMed]
- Hajjaj N, Abdulelah M, Alsharif NM, et al. Synchronous Endobronchial Carcinoid Tumor and Adenocarcinoma of the Lung: A Case Report and Review of the Literature. Cureus 2021;13:e15977. [Crossref] [PubMed]
- Shao J, Wang C, Li J, et al. A comprehensive algorithm to distinguish between MPLC and IPM in multiple lung tumors patients. Ann Transl Med 2020;8:1137. [Crossref] [PubMed]
- Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res 2012;72:4875-82. [Crossref] [PubMed]
- Murphy SJ, Aubry MC, Harris FR, et al. Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. J Clin Oncol 2014;32:4050-8. [Crossref] [PubMed]
- Schneider F, Derrick V, Davison JM, et al. Morphological and molecular approach to synchronous non-small cell lung carcinomas: impact on staging. Mod Pathol 2016;29:735-42. [Crossref] [PubMed]
- Mansuet-Lupo A, Barritault M, Alifano M, et al. Proposal for a Combined Histomolecular Algorithm to Distinguish Multiple Primary Adenocarcinomas from Intrapulmonary Metastasis in Patients with Multiple Lung Tumors. J Thorac Oncol 2019;14:844-56. [Crossref] [PubMed]
- Liu C, Liu C, Zou X, et al. Next-generation sequencing facilitates differentiating between multiple primary lung cancer and intrapulmonary metastasis: a case series. Diagn Pathol 2021;16:21. [Crossref] [PubMed]
- Zheng R, Shen Q, Mardekian S, et al. Molecular profiling of key driver genes improves staging accuracy in multifocal non-small cell lung cancer. J Thorac Cardiovasc Surg 2020;160:e71-9. [Crossref] [PubMed]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013:1303.3997.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Yen JL, Garcia S, Montana A, et al. A variant by any name: quantifying annotation discordance across tools and clinical databases. Genome Med 2017;9:7. [Crossref] [PubMed]
- Talevich E, Shain AH, Botton T, et al. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12:e1004873. [Crossref] [PubMed]
- Rausch T, Zichner T, Schlattl A, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012;28:i333-9. [Crossref] [PubMed]
- Murugaesu N, Wilson GA, Birkbak NJ, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 2015;5:821-31. [Crossref] [PubMed]
- Wang C, Sandhu J, Fakih M. A case of class 3 MEK1 mutated metastatic colorectal cancer with a non-durable tumor marker response to MEK and ERK inhibitors. J Gastrointest Oncol 2019;10:1140-3. [Crossref] [PubMed]
- Bu R, Siraj AK, Masoodi T, et al. Recurrent Somatic MAP2K1 Mutations in Papillary Thyroid Cancer and Colorectal Cancer. Front Oncol 2021;11:670423. [Crossref] [PubMed]
- Virzì AR, Gentile A, Benvenuti S, et al. Reviving oncogenic addiction to MET bypassed by BRAF (G469A) mutation. Proc Natl Acad Sci U S A 2018;115:10058-63. [Crossref] [PubMed]
- Vazquez M, Carter D, Brambilla E, et al. Solitary and multiple resected adenocarcinomas after CT screening for lung cancer: histopathologic features and their prognostic implications. Lung Cancer 2009;64:148-54. [Crossref] [PubMed]
- Wang X, Wang M, MacLennan GT, et al. Evidence for common clonal origin of multifocal lung cancers. J Natl Cancer Inst 2009;101:560-70. [Crossref] [PubMed]
- Stella F, Luciano G, Dell'Amore A, et al. Pulmonary Metastases from NSCLC and MPLC (Multiple Primary Lung Cancers): Management and Outcome in a Single Centre Experience. Heart Lung Circ 2016;25:191-5. [Crossref] [PubMed]
- Liu Y, Zhang J, Li L, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun 2016;7:13200. [Crossref] [PubMed]
- Li H, Dong S, Zhang D, et al. Targeted Sequencing Facilitated Diagnosis of an Uncommon Patient Harboring Both Multiple Primary and Intrapulmonary Metastatic Lung Cancer: A Case Report. Onco Targets Ther 2021;14:3455-9. [Crossref] [PubMed]
- Lee H, Park JH, Han J, et al. The High Proportion of Discordant EGFR Mutations among Multiple Lung Tumors. Cancers (Basel) 2022;14:3011. [Crossref] [PubMed]
- Ezer N, Wang H, Corredor AG, et al. Integrating NGS-derived mutational profiling in the diagnosis of multiple lung adenocarcinomas. Cancer Treat Res Commun 2021;29:100484. [Crossref] [PubMed]
- Zhang X, Fan X, Sun C, et al. A novel NGS-based diagnostic algorithm for classifying multifocal lung adenocarcinomas in pN0M0 patients. J Pathol Clin Res 2023;9:108-20. [Crossref] [PubMed]
- Yang CY, Yeh YC, Wang LC, et al. Genomic Profiling With Large-Scale Next-Generation Sequencing Panels Distinguishes Separate Primary Lung Adenocarcinomas From Intrapulmonary Metastases. Mod Pathol 2023;36:100047. [Crossref] [PubMed]
- Zeng C, Zhou Y, Ye W, et al. Exploration and validation of hub genes in lung adenocarcinoma based on bioinformatics analysis. Transl Cancer Res 2022;11:3814-26. [Crossref] [PubMed]
- Nicholson AG, Torkko K, Viola P, et al. Interobserver Variation among Pathologists and Refinement of Criteria in Distinguishing Separate Primary Tumors from Intrapulmonary Metastases in Lung. J Thorac Oncol 2018;13:205-17. [Crossref] [PubMed]
(English Language Editor: J. Jones)


