
Clinical and prognostic significance of detecting CEA, EGFR, LunX, c-met and EpCAM mRNA-positive cells in the peripheral blood, tumor-draining blood and bone marrow of non-small cell lung cancer patients
Highlight box
Key findings
• CTCs/DTCs play a negative prognostic role in non-metastatic NSCLC associated with poor overall, disease-free and cancer specific survival.
What is known and what is new?
• CTCs/DTCs has been identified as a negative prognostic factor in advanced NSCLC
• Bone marrow harboring DTCs represent an important reservoir for tumor cells dissemination.
What is the implication, and what should change now?
• CTCs/DTCs are eminent target for systemic therapy and disease monitoring.
Introduction
Cancer morbidity and mortality is one of the most significant problems for modern medicine. According to the latest available estimates, lung cancer ranks first among initially detected malignancies in terms of mortality, causing 13.0% and 16.7% of deaths in the general and male groups, respectively (1,2). Furthermore, lung cancer causes the largest proportion of mortality among malignant neoplasms, as 19.4% of mortalities (1.59 million from a total of 8.2 million) were attributed to lung cancer (1).
Surgical resection, which is performed in approximately 88% of non-small cell lung cancer (NSCLC) patients, can provide long-term survival (3). Nevertheless, high mortality rates remain a significant challenge despite recent advances in surgical techniques, radio-chemotherapy, targeted therapies, and the early diagnosis of NSCLC (4). Even when NSCLC is detected at an early stage, there is a 50% probability of recurrence after surgery (5,6), and the overall five-year survival rate of all stages is only 15% (7). Early metastasis, which often goes unnoticed during the initial diagnosis, further increases the mortality rate (8). Pathological tumor-nodes-metastases (TNM) classification is currently the best prognostic factor for NSCLC, but obviously this method is not perfect. Even after radical cancer surgery, metastatic potential can remain, for example, in the form of minimal residual disease (MRD). Circulating and disseminated tumor cells (CTCs and DTCs, respectively) shed from the primary tumor, enter circulation and can potentially settle into secondary organs (9,10). Circulating tumor cells were widely introduced in clinical research about ten years ago (11), and were praised as a new tumor biomarker that could be used for prognostic and predictive purposes among different types of cancer (12-14). Preoperative detection of CTCs may be useful for selecting the correct therapeutic strategy, yet their clinical significance has not yet been confirmed well. Consistently with data indicating significant prognostic role of CTCs in disease outcome, clinical trials demonstrated survival benefit of adjuvant chemotherapy in NSCLC patients (15).
CTCs of epithelial origin are present in minute amounts in the blood of patients with various forms of solid cancers, usually occurring at a rate of one per millions of normal blood cells (16). CTCs share certain characteristics with the primary tumor (17), but can become a phenotypically heterogeneous group (13,18) once they enter circulation, with certain cells undergoing epithelial-mesenchymal transition (EMT).
Recent studies indicate, the tumor metastasis is highly inefficient process. One million cells from each gram of tumor tissue enter the bloodstream daily, yet research has shown that only 0.01% of these cells will become hematogenous metastases (19,20). Over the last decade, many methods have been developed to identify, quantify and describe CTCs. Furthermore, there is ample evidence that the amount of CTCs can be regarded as an independent prognostic parameter of highly specific cancer progression in patients with breast, prostate or colon cancer (21). The American Food and Drug Administration (FDA) has only approved two methods: CellSearch for CTC enumeration and the subsequent prediction of outcome for metastatic breast, colorectal and prostate cancers (22) and Parsortix® PC1 System for the capturing and harvesting of CTCs in metastatic breast cancer patient (23). It was also proven, that CTCs presence is an independent prognostic factor of progression-free survival and overall survival in advanced NSCLC (24,25). However, the role of CTCs in early stages NSCLC progression and outcome has been much less studied, and the significance of CTCs for early lung cancer has not yet been determined.
The presented research aimed to elucidate the role of CTCs/DTCs during early stages of lung cancer. We hypothesized that the detection of CTCs or DTCs from either the proximal tumor draining (pulmonary) vein or bone marrow would signify a higher risk of post-surgery disease recurrence. To verify this hypothesis and provide information about the role of CTCs/DTCs at early stages of lung cancer, we quantified the CTC/DTC amounts in peripheral blood, pulmonary blood, and bone marrow samples of non-metastatic NSCLC patients. We present this article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-801/rc).
Methods
Patients and sample collection
The study was approved by the Institutional Review Board of the University Hospital Olomouc and the Faculty of Medicine and Dentistry (IRB number 172/08) and all participants signed an informed consent form. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was registered at ClinicalTrials.gov (NS10285). A total of 166 patients were prospectively enrolled in the study, which was conducted between August 2009 and April 2013 (26). However, 47 patients were excluded (R>0, histologically not NSCLC, stage IIIB–IV, neoadjuvant chemotherapy, death caused by perioperative complications), finally 119 patients were analyzed (Table 1).
Table 1
| Clinical stage | Parameter unit | IA | IB | IIA | IIB | IIIA | Total |
|---|---|---|---|---|---|---|---|
| Sex | N | 37 | 27 | 19 | 20 | 16 | 119 |
| Female/male | 14/23 | 6/21 | 5/14 | 6/14 | 3/13 | 34/85 | |
| Age | Years [quartiles] | 67 [62, 72] | 68 [65, 70] | 66 [60.5, 70] | 67 [60.5, 69] | 65 [57, 69] | 67 [61, 70] |
| OS (months) | N | 37 | 27 | 19 | 20 | 16 | 119 |
| FU: q50 (q25–q75) | 47 (40.25–63.9) | 53.9 (26.12–63.25) | 44 (27.7–51.48) | 12.9 (9.94–33.08) | 18.6 (13.81–24.43) | 41.6 (15.82–57.18) | |
| Events (%) | 8 (21.6) | 9 (33.3) | 7 (36.8) | 16 (80.0) | 14 (87.5) | 54 (45.4) | |
| Median (95% CI) | NA (NA, NA) | NA (NA, NA) | 65.7 (38.77, NA) | 12.9 (10.61,34.83) | 18.6 (15.24,38.9) | NA (38.77, NA) | |
| 3-y survival ± SE (%) | 83.7±6.1 | 70.4±8.8 | 73.7±10.1 | 20±8.9 | 18.8±9.8 | 59.6±4.5 | |
| CSS (months) | N | 35 | 26 | 19 | 19 | 14 | 113 |
| FU: q50 (q25–q75) | 47 (40.25–63.9) | 53.9 (26.12–63.25) | 44 (27.7–51.48) | 12.9 (9.94–33.08) | 18.6 (13.81–24.43) | 41.6 (15.82–57.18) | |
| Events (%) | 1 (2.9) | 6 (23.1) | 6 (31.6) | 9 (47.4) | 10 (71.4) | 32 (28.3) | |
| Median (95% CI) | NA (NA, NA) | NA (NA, NA) | 65.7 (NA, NA) | 20.7 (11.07, NA) | 21.2 (15.24, NA) | NA (NA, NA) | |
| 3-y survival ± SE (%) | 96.9±3.1 | 80.6±7.8 | 73.7±10.1 | 39.9±13.6 | 27.1±13 | 72.6±4.4 | |
| DFS (months) | N | 37 | 27 | 19 | 20 | 16 | 119 |
| FU: q50 (q25–q75) | 40.8 (24.02–45.8) | 41 (12.63–53.45) | 15.4 (8.85–35.81) | 11 (5.62–28.49) | 11.7 (5.45–16.37) | 27.2 (10.19–43.37) | |
| Events (%) | 7 (18.9) | 6 (22.2) | 9 (47.4) | 10 (50.0) | 10 (62.5) | 42 (35.3) | |
| Median (95% CI) | NA (47.18, NA) | NA (NA, NA) | 38.8 (18.5, NA) | 15.8 (10.32, NA) | 13.4 (11.7, NA) | NA (41.56, NA) | |
| 3-y survival ± SE (%) | 89.1±6 | 80±8 | 53.9±13.5 | 35.8±13.1 | 11.5±10.6 | 64.6±4.9 | |
| Histology | AC | 25/37 (67.6) | 12/27 (44.4) | 8/19 (42.1) | 7/20 (35.0) | 5/16 (31.2) | 57/119 (47.9) |
| NAC | 12/37 (32.4) | 15/27 (55.6) | 11/19 (57.9) | 13/20 (65.0) | 11/16 (68.8) | 62/119 (52.1) | |
| Grading | 1 | 6/37 (16.2) | 1/27 (3.7) | 2/19 (10.5) | 1/20 (5.0) | 0/16 (0) | 10/119 (8.4) |
| 2 | 18/37 (48.6) | 3/27 (11.1) | 4/19 (21.1) | 8/20 (40.0) | 6/16 (37.5) | 39/119 (32.8) | |
| 3 | 13/37 (35.1) | 23/27 (85.2) | 13/19 (68.4) | 11/20 (55.0) | 10/16 (62.5) | 70/119 (58.8) | |
| T | 1 | 37/37 (100.0) | 1/27 (3.7) | 3/19 (15.8) | 1/20 (5.0) | 3/16 (18.8) | 45/119 (37.8) |
| 2 | 0/37 (0) | 26/27 (96.3) | 15/19 (78.9) | 15/20 (75.0) | 3/16 (18.8) | 59/119 (49.6) | |
| 3 | 0/37 (0) | 0/27 (0) | 1/19 (5.3) | 4/20 (20.0) | 10/16 (62.5) | 15/119 (12.6) | |
| N | 0 | 37/37 (100.0) | 27/27 (100.0) | 8/19 (42.1) | 4/20 (20.0) | 0/16 (0) | 76/119 (63.9) |
| 1+2 | 0/37 (0) | 0/27 (0) | 11/19 (57.9) | 16/20 (80.0) | 16/16 (100.0) | 43/119 (36.1) | |
| Treatment | RT/total (%) | 0/37 (0) | 0/27 (0) | 0/19 (0) | 1/20 (5.0) | 3/16 (18.8) | 4/119 (3.4) |
| ChT/total (%) | 3/37 (8.1) | 19/27 (70.4) | 14/19 (73.7) | 16/20 (80.0) | 16/16 (100.0) | 68/119 (57.1) |
OS, overall survival; CSS, cancer-specific survival; DFS, disease-free survival; T, tumor category from TNM classification; N, node category from TNM classification; FU, follow-up; AC, adenocarcinoma; NAC, non-adenocarcinoma; RT, radiotherapy; ChT, adjuvant chemotherapy; y, years; SE, standard error; CI, confidence interval; NA, not available.
Peripheral blood (PB) (from the cubital vein before surgery), tumor-draining pulmonary vein blood (TDB) (from a major tumor-draining vein immediately before clamping), bone marrow (BM) (by sternal puncture after the anesthesia induction) and tumor tissue samples (during surgery) were collected from the patients. Samples was drawn into vacutainer tubes containing 200 µL of 10% EDTA (BD, Franklin Lakes, NJ, USA), and immediately transported to the laboratory for further processing. To prevent any epithelial cell contamination during sampling, the first 2–3 mL of peripheral blood was discarded, after which the remainder was analyzed. The average time from blood sampling to processing did not exceed two hours. Fresh tumor tissue samples were stored in RNAlater® solution (Qiagen, Valencia, CA, USA) and as formalin-fixed paraffin embedded (FFPE) tumor tissues.
Detection of K-ras, BRAF and EGFR mutations
The DNA was purified from FFPE tumor tissue using cobas® DNA Sample Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany) concurring with standard device settings and conditions and according to the manufacturer’s instructions. Mutations of the Kirsten rat sarcoma viral oncogene homolog (K-ras) in tumor genomic DNA were detected using TheraScreen KRAS RGQ PCR (DxS, Manchester, UK). The TheraScreen kit can detect the seven most common K-ras mutations at codons 12 and 13 (G12A, G12V, G12C, G12D, G12S, G12R, G13D) (27). In the PRIME trial some additional predictive RAS gene mutations (KRAS codons 59, 61, 117 and 146; NRAS codons 12 and 13) have been identified later (28) (but these mutations were not tested in the present study. Mutations of the murine sarcoma viral oncogene homolog B (BRAF) in tumor genomic DNA were detected using the BRAF p.Val600Glu kit (IntellMed, Olomouc, Czech Republic). The kit can detect the most common BRAF mutation, V600E. Mutations of the epidermal growth factor receptor (EGFR) in tumor genomic DNA were detected using cobas® EGFR Mutation Test (Roche Diagnostics, Basel, Switzerland). The kit can detect the most frequent mutations in lung cancer at exon 18 (G719X, G719A, G719C, G719S), 29 different deletions at exon 19, the S768I and T790M mutations as well as 5 different insertions at exon 20, and the L858R mutation at exon 21.
Fluorescence in situ hybridization analysis (FISH)
FISH analysis was performed on FFPE tissues according to the manufacturer’s protocol. We used break-apart probes for ALK (anaplastic lymphoma kinase) (Cytocell Ltd., Cambridge, United Kingdom) and ROS1 (ros protooncogene 1) (IntellMed, Olomouc, Czech Republic) gene rearrangements, along with LSI c-myc (myc protooncogen)/CEP8, LSI FGFR1 (fibroblast growth factor receptor 1)/CEP8, LSI EGFR/CEP7 and LSI c-met (met protooncogen)/CEP7 (all provided by IntellMed, Olomouc, Czech Republic) probes for gene/chromosome copy number enumeration. The signals were observed and quantified using fluorescence microscopy. At least 100 non-overlapping nuclei were selected in each sample. FISH positivity for ROS1 and ALK rearrangement was defined as split (ROS1, ALK) or single red (ALK) signal in >15% of the nuclei. Increased, or decreased, copy number was defined as >30% of the nuclei showing a ≥3 or <2-fold-change, respectively, in signal, or when the average signal exceeded the cut-off limits. Limits of >2.5 and <1.8 copy number/nucleus were set for subsequent statistical analyses (29).
RNA purification and reverse transcription
The total white blood cells were isolated by osmotic lysis from peripheral blood and bone marrow samples. Briefly, 10 mL of blood was lysed in 40 mL of hypotonic lysis buffer containing 1.55 M NH4Cl, 0.1 M NH4HCO3 and 1 mM EDTA for 15–40 minutes on ice. Aliquots of 1.1×107 cells were resuspended in 1 mL of TRIreagent (Molecular Research Center, Cincinnati, USA) and total RNA was extracted according to manufacturer’s instructions. Prior to RNA purification, tumor tissue samples (30–50 mg) in RNAlater® (Qiagen, Hilden, Germany) were homogenized in 1 mL of Trizol using a MixerMill 301 homogenizer (Retsch, Haan, Germany). RNA concentration and purity were assessed using a Nanodrop ND 1000 instrument (ThermoScientific, Wilmington, DE, USA). Reverse transcription was performed on 3 µg of total RNA using random primers (Promega, Madison, Wisconsin, USA), RNAsin ribonuclease inhibitor (Promega, Madison, Wisconsin, USA), and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) in a 30 µL reaction volume according to the manufacturer’s instructions (30). Samples were stored at −20 °C until qPCR analysis.
Primer and probe design
Primers and probes for the amplification of CEA, EGFR, LunX (lung specific protein X, resp. BPIFA1—BPI fold-containing protein A1), c-met and EpCAM cDNA were designed using the PrimerPremier3 software (Premier Biosoft, Palo Alto, CA, USA) and NCBI Reference sequences (accession numbers NM 004363, NM 005228, NM 016583, NM 000245 and NM 002354, respectively). Each primer set spanned an intron sequence to prevent genomic DNA amplification: CEA-fw 5'-taagtgttgaccacagcgaccc-3', CEA-rev 5'-gttcccatcaatcagccaagaa-3' and CEA-probe 5'-atgtcctctatggcccagacgaccc-3' BHQ1-HEX (167 bp amplicon); EGFR-fw 5'-acttcaaaaactgcacctccat-3', EGFR-rev 5'-aatcagcaaaaaccctgtgatt-3' and EGFR-probe 5'-acatcctgccggtggcatttagg-3' BHQ1-HEX (149 bp amplicon); LunX-fw 5'-gatggccaccgtctctatgt-3', LunX-rev 5'-acagccagcctcaacagact-3' and LunX-probe 5'-ccatccctctcggcataaagctcc-3' BHQ1-HEX (93 bp amplicon); c-met-fw 5'-tggacaatgatggcaagaaa-3', c-met-rev 5'-gatgattccctcggtcagaa-3' and c-met-probe 5'-tcactgtgctgtgaaatccttgaaca-3' BHQ1-HEX (99 bp amplicon); EpCAM-fw 5'-aaacacaaagcaagagaaaaacct-3', EpCAM-rev 5'-aattttggatccagttgataacg-3' and EpCAM-probe 5'-ttgcggactgcacttcagaagga-3' BHQ1-HEX (95 bp amplicon) (Generi-Biotech, Hradec Kralove, Czech Republic) (31).
Quantitative real-time polymerase chain reaction
The amount of CEA, EGFR, LunX, c-met and EpCAM mRNA was quantified through quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). A reaction volume of 25 µL was used, and this contained: 1 U of HotStart Taq Polymerase; 10× PCR buffer (AB Gene, Epsom, UK); 200 µM dNTPs (Promega), an optimized amount of primers (primer-Fw—300 nM for CEA and 400 nM for EGFR, LunX, c-met, EpCAM; primer-Rev—600 nM for CEA and 400 nM for EGFR, LunX, c-met, EpCAM); probes (200 nM for each); magnesium cations (3 mM for CEA and EGFR; 4 mM for LunX; 6 mM for c-met and EpCAM); and 100 ng of cDNA. Reaction progress was followed on a Rotor Gene 3000 (Corbett Research, Sydney, Australia). The optimized thermal profile for amplification consisted of an initial polymerase activation at 96 °C for 15 minutes followed by 50 cycles of: 95 °C for 15 seconds and then either 65 °C for 15 seconds (CEA), 62 °C for 15 seconds (EGFR), 62 °C for 10 seconds (LunX), 58 °C for 15 seconds (c-met), or 59 °C for 15 seconds (EpCAM).
Standardization and normalization of the data
Standard curves were constructed to determine the exact number of mRNA copies for each gene that was tested. The standards were generated by using outer primers for each marker to amplify gene-specific sequences. The principle of nested PCR was used to analyze amplicons together with the samples. Absolute mRNA copy numbers for each gene were calculated using concentrations measured on NanoDrop ND 1000 (Thermo Fisher Scientific, Waltham, MA, USA), the molecular weight of the amplicon, and Avogadro’s constant (6.023×1023 mol). In order to avoid intra- and inter-individual variability of reference gene expression, CEA, EGFR, LunX, c-met and EpCAM expression was normalized to the amount of total RNA used in each reverse transcription reaction. The qRT-PCR setup followed MIQE guidelines (32).
Method verification and sensitivity
The experimental samples containing human buffy coat cells and the lung adenocarcinoma cell line HCI-H2228 (ATCC, Manassa, VA, USA; RRID:CVCL_1543) were prepared to validate the sensitivity and specificity of the qRT-PCR method used in the presented study. The total numbers of 10, 20, 50, 100, 200, 500, 1,000 and 2,000 HCI-H2228 cells were directly sorted based on the forward and side scatter parameters (FACS Aria, Beckman, Indianapolis, USA) into white blood cell aliquots (10 million buffy coat cells). These experimental samples were tested using qRT-PCR according to the same conditions as described above. In parallel, identical samples were analyzed using colony formation assay (CFA).
For CFA, the experimental samples were resuspended with buffy coat cells in RPMI 1640 medium containing 10% fetal calf serum and antibiotics. The dilution mixtures were cultivated in six-well flat bottom plates (TPP, Trasadingen, Switzerland) in a humidified 5% CO2 chamber at 37 °C. After one day of incubation, unattached buffy coat cells were washed away with 1% phosphate buffer saline (PBS) and the attached cells were cultured in RPMI 1640 medium. After 13 days of incubation, the colonies were washed with 1% PBS, fixed with 1% paraformaldehyde, stained with crystal violet and counted.
Statistical analysis
All of the statistical analyses were carried out in R software (www.r-project.org), ver. 4.1.0 including the following additional R packages: exactRankTests, ver. 0-8.34, dplyr, ver. 1.0.8 and maxstat, ver. 0-7.25 (33). Kruskal-Wallis/ANOVA tests and Wilcoxon exact rank/Student’s t-tests were used to evaluate differences in various clinical and pathological variables between the experimental groups. Contingency table analysis was performed to evaluate whether various clinical and pathological characteristics were dependent on CTC/DTC presence. The Bonferroni correction was applied to correct for multiple comparisons. The Kaplan-Meier method and log-rank tests were used to assess whether the detection of CEA, EGFR, LunX, c-met and EpCAM mRNA-positive cells in blood and bone samples influenced DFS, CSS and overall OS survival rates. The same method was used to assess how c-myc, ROS1, ALK, EGFR, c-met, FGFR1, K-ras, and BRAF mutations affect patient survival, as well as other clinical and pathological parameters. The Cox regression univariate analysis was also performed, and results are presented as a hazard ratio (HR) with a 95% confidential interval (CI). Adjusted Cox regression models for CEA, EGFR, LunX, c-met and EpCAM mRNA-positive cells (adjusted to age at diagnosis, gender and cancer stage) in the blood (peripheral and pulmonary) and bone marrow were analyzed. Multivariate Cox regression models for OS, DFS, and CSS were built and the independent variables were selected by stepwise selection from CEA and EpCAM mRNA in blood and bone marrow (stage, age and gender were included in all models as adjustment factors). The significance threshold was set at P=0.05.
CEA, EGFR, LunX, c-met and EpCAM mRNA gene expression was analyzed and is presented on two categorical levels: negative and positive values. Cut-off values for each parameter (CEA, EGFR, LunX, c-met, EpCAM) and compartment (peripheral blood, pulmonary blood and bone marrow) were set according to disease-free survival (Figure 1). An initial cut-off estimation for each marker and compartment was established through the maxstat() function in R package; this provided an estimate that had been obtained from 10,000 randomly selected samples chosen from a dataset of 117 patients (two patients with follow-up <3 years and without DFS event were excluded) with stage I, II or III and a DFS event three years after surgery or a follow-up of at least three years. The raw data are accessible at https://doi.org/10.6084/m9.figshare.11920452.v1.
The ratio of patients with and without a DFS event in each random sample group was equal to the initial ratio, and each of the random samples contained at least 90% of all the 117 patients. Next, the minimum of cut-offs for each marker and compartment was calculated. The variables included in the multivariate analysis were stage, age and gender of patients, stage of the disease, tumor size and presence/absence of metastasis in regional lymph nodes, histological characteristics such as tumor type and malignancy, tobacco-smoking status and presence of CEA-, EGFR-, Lunx-, c-met-, and EpCAM-positive CTCs/DTCs. Important prognostic criteria were established using Bayesian Model Averaging (BMA). Multivariate Cox regression models were then built from the selected criteria.
Results
Verification of qRT-PCR method
The qRT-PCR method used in the present study was compared to the CFA method to determine the sensitivity of CTCs detection. The HCI-H2228 colonies were identified in all experimental samples using CFA. Similarly, the CEA and EpCAM gene expression was higher than the cut-off values calculated for clinical samples (Table 2) in all tested experimental samples using qRT-PCR (Figure 2). Both the qRT-PCR and CFA methods reproducibly identified 10 HCI-H2228 cells in 10 million buffy coat cells. The comparison confirmed a statistically significant correlation among the results obtained by both, CEA and EpCAM mRNA expression versus CFA method (for both r>0.95, P≤0.001).
Table 2
| Characteristics | CEA [positive/total (percentage)] | EGFR [positive/total (percentage)] | LunX [positive/total (percentage)] | c-met [positive/total (percentage)] | EpCAM [positive/total (percentage)] | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TDB | PB | BM | TDB | PB | BM | TDB | PB | BM | TDB | PB | BM | TDB | PB | BM | |||||
| Cut-off values (gene copies/μg RNA) | 390 | 330 | 485 | 455 | 335 | 46,190 | 150 | 65 | 400 | 11,970 | 6,250 | 5,520 | 3,677,540 | 1,291,500 | 20,572,400 | ||||
| Sex | |||||||||||||||||||
| F | 9/31 (29.0) | 10/34 (29.4) | 11/33 (33.3) | 12/31 (38.7) | 9/34 (26.5) | 10/33 (30.3) | 5/31 (16.1) | 7/34 (20.6) | 13/33 (39.4) | 2/31 (6.5) | 8/34 (23.5) | 20/33 (60.6) | 9/31 (29.0) | 9/34 (26.5) | 12/33 (36.4) | ||||
| M | 25/81 (30.9) | 21/84 (25.0) | 38/82 (46.3) | 19/81 (23.5) | 12/84 (14.3) | 25/82 (30.5) | 8/81 (9.9) | 25/84 (29.8) | 27/82 (32.9) | 12/81 (14.8) | 17/84 (20.2) | 37/82 (45.1) | 16/81 (19.8) | 23/84 (27.4) | 37/82 (45.1) | ||||
| Clinical stage | |||||||||||||||||||
| IA | 7/32 (21.9) | 11/37 (29.7) | 16/35 (45.7) | 9/32 (28.1) | 5/37 (13.5) | 10/35 (28.6) | 4/32 (12.5) | 13/37 (35.1) | 15/35 (42.9) | 4/32 (12.5) | 5/37 (13.5) | 17/35 (48.6) | 7/32 (21.9) | 10/37 (27.0) | 16/35 (45.7) | ||||
| IB | 10/26 (38.5) | 7/27 (25.9) | 7/26 (26.9) | 7/26 (26.9) | 3/27 (11.1) | 10/26 (38.5) | 4/26 (15.4) | 6/27 (22.2) | 10/26 (38.5) | 1/26 (3.8) | 5/27 (18.5) | 14/26 (53.8) | 5/26 (19.2) | 6/27 (22.2) | 8/26 (30.8) | ||||
| IIA | 3/19 (15.8) | 5/19 (26.3) | 9/19 (47.4) | 8/19 (42.1) | 7/19 (36.8) | 5/19 (26.3) | 4/19 (21.1) | 8/19 (42.1) | 4/19 (21.1) | 4/19 (21.1) | 4/19 (21.1) | 13/19 (68.4) | 4/19 (21.1) | 5/19 (26.3) | 10/19 (52.6) | ||||
| IIB | 6/19 (31.6) | 3/19 (15.8) | 8/19 (42.1) | 3/19 (15.8) | 3/19 (15.8) | 7/19 (36.8) | 0/19 (0) | 1/19 (5.3) | 4/19 (21.1) | 1/19 (5.3) | 6/19 (31.6) | 8/19 (42.1) | 2/19 (10.5) | 3/19 (15.8) | 10/19 (52.6) | ||||
| IIIA | 8/16 (50.0) | 5/16 (31.2) | 9/16 (56.2) | 4/16 (25.0) | 3/16 (18.8) | 3/16 (18.8) | 1/16 (6.2) | 4/16 (25.0) | 7/16 (43.8) | 4/16 (25.0) | 5/16 (31.2) | 5/16 (31.2) | 7/16 (43.8) | 8/16 (50.0) | 5/16 (31.2) | ||||
| Grading | |||||||||||||||||||
| G1 | 2/10 (20.0) | 4/10 (40.0) | 4/9 (44.4) | 3/10 (30.0) | 0/10 (0) | 2/9 (22.2) | 1/10 (10.0) | 4/10 (40.0) | 4/9 (44.4) | 2/10 (20.0) | 3/10 (30.0) | 6/9 (66.7) | 2/10 (20.0) | 3/10 (30.0) | 5/9 (55.6) | ||||
| G2 | 9/34 (26.5) | 5/38 (13.2) | 16/37 (43.2) | 9/34 (26.5) | 4/38 (10.5) | 14/37 (37.8) | 3/34 (8.8) | 10/38 (26.3) | 15/37 (40.5) | 5/34 (14.7) | 10/38 (26.3) | 22/37 (59.5) | 8/34 (23.5) | 10/38 (26.3) | 16/37 (43.2) | ||||
| G3 | 23/68 (33.8) | 22/70 (31.4) | 29/69 (42.0) | 19/68 (27.9) | 17/70 (24.3) | 19/69 (27.5) | 9/68 (13.2) | 18/70 (25.7) | 21/69 (30.4) | 7/68 (10.3) | 12/70 (17.1) | 29/69 (42.0) | 15/68 (22.1) | 19/70 (27.1) | 28/69 (40.6) | ||||
| T | |||||||||||||||||||
| 1 | 9/40 (22.5) | 11/45 (24.4) | 20/43 (46.5) | 13/40 (32.5) | 7/45 (15.6) | 14/43 (32.6) | 6/40 (15.0) | 16/45 (35.6) | 18/43 (41.9) | 6/40 (15.0) | 6/45 (13.3) | 21/43 (48.8) | 8/40 (20.0) | 12/45 (26.7) | 21/43 (48.8) | ||||
| 2 | 20/57 (35.1) | 15/58 (25.9) | 21/57 (36.8) | 14/57 (24.6) | 10/58 (17.2) | 18/57 (31.6) | 6/57 (10.5) | 12/58 (20.7) | 16/57 (28.1) | 6/57 (10.5) | 15/58 (25.9) | 31/57 (54.4) | 11/57 (19.3) | 13/58 (22.4) | 24/57 (42.1) | ||||
| 3+4 | 5/15 (33.3) | 5/15 (33.3) | 8/15 (53.3) | 4/15 (26.7) | 4/15 (26.7) | 3/15 (20.0) | 1/15 (6.7) | 4/15 (26.7) | 6/15 (40.0) | 2/15 (13.3) | 4/15 (26.7) | 5/15 (33.3) | 6/15 (40.0) | 7/15 (46.7) | 4/15 (26.7) | ||||
| N | |||||||||||||||||||
| 0 | 18/70 (25.7) | 21/76 (27.6) | 28/73 (38.4) | 18/70 (25.7) | 12/76 (15.8) | 23/73 (31.5) | 9/70 (12.9) | 23/76 (30.3) | 27/73 (37.0) | 7/70 (10.0) | 13/76 (17.1) | 37/73 (50.7) | 13/70 (18.6) | 17/76 (22.4) | 32/73 (43.8) | ||||
| 1+2 | 16/42 (38.1) | 10/42 (23.9) | 21/42 (50.0) | 13/42 (30.9) | 9/42 (21.4) | 12/42 (28.6) | 4/42 (9.5) | 9/42 (21.4) | 13/42 (30.9) | 7/42 (16.7) | 12/42 (28.6) | 20/42 (47.6) | 12/42 (28.6) | 15/42 (35.7) | 17/42 (40.5) | ||||
| Smoking | |||||||||||||||||||
| No | 4/12 (33.3) | 4/12 (33.3) | 6/12 (50.0) | 4/12 (33.3) | 6/12 (50.0) | 3/12 (25.0) | 0/12 (0) | 4/12 (33.3) | 6/12 (50.0) | 1/12 (8.3) | 3/12 (25.0) | 8/12 (66.7) | 3/12 (25.0) | 2/12 (16.7) | 6/12 (50.0) | ||||
| Ex | 12/40 (30.0) | 9/41 (22.0) | 18/41 (43.9) | 9/40 (22.5) | 6/41 (14.6) | 11/41 (26.8) | 7/40 (17.5) | 11/41 (26.8) | 16/41 (39.0) | 5/40 (12.5) | 10/41 (24.4) | 20/41 (48.8) | 8/40 (20.0) | 10/41 (24.4) | 22/41 (53.7) | ||||
| Yes | 15/51 (29.4) | 16/56 (28.6) | 22/53 (41.5) | 15/51 (29.4) | 8/56 (14.3) | 20/53 (37.7) | 5/51 (9.8) | 17/56 (30.4) | 17/53 (32.1) | 7/51 (13.7) | 12/56 (21.4) | 24/53 (45.3) | 10/51 (19.6) | 14/56 (25.0) | 15/53 (28.3) | ||||
| Age (years), median (q25–q75) | |||||||||||||||||||
| CTCs negative | 66 [59–70] | 66 [60–70] | 67 [61–70] | 67 [61–70] | 67 [61–70] | 66 [59–70] | 67 [61.5–70] | 66.5 [61–70] | 67 [62.5–70.5] | 67 [61–70] | 67 [61–70] | 67 [61.25–70] | 66 [60.5–70] | 66 [60–70] | 66 [60.25–70] | ||||
| CTCs positive | 68.5 [65.25–70] | 69 [65.5–70] | 67 [62–72] | 67 [63.5–70] | 67 [60–69] | 67 [66–70] | 66 [60–69] | 68 [64.25–70.25] | 66.5 [60–70] | 66 [65–70] | 66 [62–70] | 67 [60–70] | 68 [66–69] | 68.5 [66–70] | 67 [65–72] | ||||
| Pos./total (%) | 34/112 (30.4) | 31/118 (26.3) | 49/115 (42.6) | 31/112 (27.7) | 21/118 (17.8) | 35/115 (30.4) | 13/112 (11.6) | 32/118 (27.1) | 40/115 (34.8) | 14/112 (12.5) | 25/118 (21.2) | 57/115 (49.6) | 25/112 (22.3) | 32/118 (27.1) | 49/115 (42.6) | ||||
NSCLC, non-small cell lung cancer; CEA, carcinoembryonic antigen; EGFR, epidermal growth factor receptor; LunX, lung specific protein X; c-met, met protooncogene; EpCAM, epithelial cellular adhesion molecule; TDB, tumor-draining blood; PB, peripheral blood; BM, bone marrow; T, tumor category from TNM classification; N, node category from TNM classification; CTCs, circulating tumor cells.

Patients’ characteristics and presence of CTCs/DTCs
In total, 119 patients with lung cancer were included in the study (Table 1). All of the patients underwent curative surgery (R0), all tumors were histologically verified as non-small cell lung cancer and none of the enrolled patients received neoadjuvant therapy. Fifty-four (45.4%) patients died in median follow-up 41.6 months. The 11.5% (7/61), 39.5% (15/38) and 71.4% (10/14) of patients stage I, II and III respectively died from lung cancer. The group included 56 smokers, 42 ex-smokers and 12 non-smokers. All 119 patients were followed and treated in accordance with standard guidelines (34). No targeted therapy or immunotherapy were administered in first line of adjuvant therapy. CTC/DTC presence, as determined by CEA, EGFR, LunX, c-met or EpCAM mRNA expression, was further categorized by gender, age, smoking status, clinical stage, T/N category and histological tumor grade (Table 2). We found no correlations between the presence of CTCs/DTCs and conventional clinical prognostic characteristics, such as the tumor size, local lymph node metastases, stage of the disease, tumor histological type and the malignancy level (Table 2). A statistically significant correlation was seen only in case of the smoking status. However, the patient population is unevenly distributed, with many more smokers (56) or ex-smokers (41) than non-smokers (12). This phenomenon is characteristic of this disease.
Tumor tissue genomic landscape
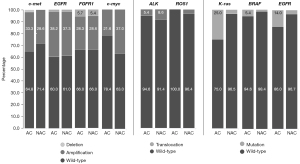
EGFR mutations were identified in 8.5% (10/118) of the tumors using PCR, and FISH detected amplifications and deletions of the gene in 37.7% (43/114) and 1.8% (2/114) of the cancers, respectively (Figure 3). The KRAS gene mutations at codons 12 and 13 were identified in 13.8% (15/109) of the tumors, more frequent in adenocarcinoma than non-adenocarcinoma (25% vs. 3.5% respectively). Amplification and deletion of the c-met gene was found in 30.9% (34/110) and 0.9% (1/110) of the tumors, respectively. The analyses showed amplification and deletion of the FGFR1 gene in 28.4% (31/109) and 5.5% (6/109) of the tumors, respectively. C-myc was amplified in 29.5% (31/105) of the tumors. ALK and ROS1 translocations were detected in 7% (8/114) and 1.9% (2/106) of the tumors, respectively (Figure 3). The described mutations, amplifications, deletions, and translocations did not correlate with survival rates.
Prognostic significance of CTCs/DTCs in survival analysis
All 119 patients were followed and treated in accordance with standard guidelines. The prognostic significance of CTCs/DTCs was assessed in specimens collected before and during the surgery. In total, 54 (45.4%) of the 119 stage IA–IIIA NSCLC patients died during the follow-up period, and 32 of these deaths were connected to the current cancer disease (Table 1). The presence of CEA mRNA-positive CTCs in tumor draining pulmonary blood samples indicated significantly shorter CSS (HR =2.5; 95% CI: 1.21–5.08; P=0.013) and CEA mRNA-positive DTCs in the bone marrow indicated significantly shorter OS (HR =2; 95% CI: 1.16–3.44; P=0.013) and CSS (HR =2.1; 95% CI: 1.04–4.3; P=0.038), when compared to patients without CEA mRNA-positive CTCs/DTCs in univariate analysis (Figure 4). The presence of CEA mRNA-positive CTCs in preoperative peripheral blood samples had nearly significant impact on DFS (HR =1.9; 95% CI: 0.99–3.5; P=0.055) and CSS (HR =2; 95% CI: 0.95–4.01; P=0.067).

The presence of EpCAM mRNA-positive CTCs in tumor draining pulmonary blood impacted CSS (HR =2.3; 95% CI: 1.08–4.79; P=0.031) and DFS (HR =2; 95% CI: 1.01–3.86; P=0.045). The other tested markers (LunX, c-met, EGFR) did not show any statistically significant effects in the univariate survival analysis. The adjusted survival analysis revealed that CEA mRNA-positive CTCs/DTCs in peripheral blood [HR(DFS) =2.75, P=0.004; HR(CSS) =2.77, P=0.009] and bone marrow [HR(OS) =1.96, P=0.024] are independent prognostic factors (Table 3).
Table 3
| Marker | Compartment | DFS | CSS | OS | |||||
|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||||
| CEA | TDB | 1.24 (0.57, 2.67) | 0.591 | 1.52 (0.65, 3.56) | 0.339 | 1.09 (0.57, 2.09) | 0.788 | ||
| PB | 2.75 (1.38, 5.47) | 0.004 | 2.77 (1.29, 5.98) | 0.009 | 1.62 (0.87, 3.01) | 0.127 | |||
| BM | 1.53 (0.79, 2.97) | 0.212 | 1.88 (0.87.4.06) | 0.110 | 1.96 (1.09, 3.53) | 0.024 | |||
| EGFR | TDB | 0.36 (0.15, 0.85) | 0.020 | 0.39 (0.14, 1.09) | 0.074 | 0.34 (0.15, 0.78) | 0.010 | ||
| PB | 1.65 (0.67, 4.07) | 0.274 | 2.49 (0.97, 6.41) | 0.058 | 1.93 (0.97, 3.83) | 0.061 | |||
| BM | 0.74 (0.36, 1.49) | 0.395 | 0.89 (0.40, 1.94) | 0.763 | 1.01 (0.57, 1.80) | 0.977 | |||
| LunX | TDB | 0.42 (1.12, 1.45) | 0.171 | 0.43 (0.10, 1.89) | 0.265 | 0.51 (0.15, 1.69) | 0.271 | ||
| PB | 0.70 (0.30, 1.62) | 0.399 | 0.90 (0.32, 2.54) | 0.837 | 1.18 (0.60, 2.29) | 0.631 | |||
| BM | 0.57 (0.27, 1.18) | 0.130 | 0.70 (0.30, 1.63) | 0.403 | 0.81 (0.44, 1.50) | 0.506 | |||
| c-met | TDB | 1.26 (0.49, 3.21) | 0.628 | 1.06 (0.37, 3.07) | 0.910 | 0.01 (0.42, 2.42) | 0.976 | ||
| PB | 0.91 (0.46, 1.83) | 0.799 | 0.61 (0.26, 1.44) | 0.262 | 0.79 (0.42, 1.48) | 0.455 | |||
| BM | 0.48 (0.24, 0.95) | 0.036 | 0.65 (0.30, 1.41) | 0.278 | 0.60 (0.34, 1.09) | 0.093 | |||
| EpCAM | TDB | 1.65 (0.82, 3.34) | 0.163 | 1.98 (0.88, 4.47) | 0.100 | 1.47 (0.76, 2.83) | 0.248 | ||
| PB | 0.96 (0.48, 1.92) | 0.908 | 1.29 (0.60, 2.75) | 0.510 | 1.00 (0.54, 1.85) | 0.989 | |||
| BM | 0.96 (0.50, 1.83) | 0.895 | 0.98 (0.48, 2.04) | 0.966 | 1.01 (0.58, 1.76) | 0.965 | |||
DFS, disease-free survival; CSS, cancer-specific survival; OS, overall survival; TDB, tumor-draining blood; PB, peripheral blood; BM, bone marrow; CEA, carcinoembryonic antigen; EGFR, epidermal growth factor receptor; LunX, lung specific protein X; c-met, met protooncogene; EpCAM, epithelial cellular adhesion molecule; HR, hazard ratio; CI, confidence interval.
Furthermore, the EGFR mRNA-positive CTCs in the tumor draining pulmonary blood was shown to significantly impact DFS and OS (HR =0.36, P=0.020; HR =0.34, P=0.010, respectively) in adjusted survival analysis.
Moreover, in a multivariate analysis (using a multivariate Cox regression model), the presence of CEA mRNA-positive CTCs/DTCs in the peripheral blood and bone marrow sampled at surgery was identified as an independent negative prognostic factor for DFS (HR =2.79, P=0.005; HR =1.96, P=0.057, respectively). As expected, higher clinical stage was an independent prognostic factor for shorter OS, CSS and DFS.
Discussion
Method of choice
CTC detection has been widely used in clinical studies during the last ten years (11). Their increasing significance in clinical research can be confirmed by the amount of research focusing on CTCs found on the registry of clinical trials, www.clinicaltrials.gov. CTCs clearly have a decisive role in the progression of cancer to metastasis and have been shown to decrease the survival rate (35,36). The various methods used for CTC detection can be divided into cell-based detection and nucleic acid-based detection (37). CellSearch was approved by FDA for CTCs detection in metastatic breast, colorectal and prostate cancers (22). There are several studies using this method in lung cancer, in which confirmed the presence of CTCs in patients with NSCLC (in 23–78% patients) (38-41) and have shown shorter PFS and OS in this patients (38,39). In this study, we used qRT-PCR method, which is highly sensitive to detect indirectly CTCs based on presence of tumor-specific mRNA in peripheral blood or bone marrow samples. Furthermore, we have proven the suitability of qRT-PCR for CTC detection by comparing it with gold standard colony-forming assay. Importantly, this method could be widely used at low cost in any molecular laboratory equipped with thermal cyclers not requiring further investments.
Sampling sites
Numerous previous studies have shown that CTCs exist in peripheral blood in minute amounts. This phenomenon is especially noticeable in lung cancer. Nevertheless, it is possible to sample CTCs at the site of their primary dissemination into the blood stream (namely from the tumor draining pulmonary vein) and their main reservoir (bone marrow), both of which are locations where the probability of detection is theoretically higher than that of peripheral blood. This has been confirmed by Okumura et al., who utilized CellSearch to demonstrate the visual difference between peripheral and pulmonary blood in terms of CTC presence (5 out of 30 patients had CTCs in peripheral blood whereas 29 out of 30 had CTCs in the pulmonary blood) (42). Similar results were described in another work (43), where 25% of patients (8 out of 32) had CTCs in peripheral blood, while 29 of 32 pulmonary blood samples was CTCs positive. In addition, Crosbie et al., also reported a more frequent detection of CTCs in the pulmonary blood than in peripheral blood (43% against 22%) and the total number of the CTCs in the study was also greater in the pulmonary vein (P=0.002) (3). In our study, we found the tumor draining pulmonary blood and bone marrow samples expressed several-fold larger amounts of mRNA of all 5 tested markers than peripheral blood samples. Patients with CEA mRNA-positive CTCs in the peripheral blood trended to have decreased CSS and DFS, but the result was not statistically significant. In contrast, the presence of CEA mRNA-positive CTCs in pulmonary blood was a significant negative predictor for CSS (HR =2.5; P=0.013). This result corresponds with the findings of a study by Sienel (2003). The authors presented a multivariate analysis that showed that the presence of CTCs in pulmonary vein tends to be linked to a poor CSS (P=0.054), and is a significant poor prognostic factor of CSS in subgroups with mediastinal lymph nodes absence (RR =4.2; 95% CI: 1.6–11.1; P=0.004) (44). In our study, the adjusted and multivariate Cox regression analyses also showed that CEA mRNA-positive CTCs presence in peripheral blood is the independent negative prognostic factor. DTCs can persist in bone marrow for a long time due to the interaction with its microenvironment (45). However, the significance of DTCs as a prognostic factor remains ambiguous. Several studies have reported that the presence of DTCs has no effect on patient survival (46-48), while another study (49) pointed out that the presence of cytokeratin-positive DTCs, detected with low sensitivity immunocytochemistry method using AE1/AE3 antibodies, was associated with DFS reduction in some patient subgroups. However, in the latter study, the presence of cytokeratin-positive cells did not show a significant prognostic impact on the whole group of patients (P=0.26). This phenomenon can be explained by the fact that some of the tumor cells found in bone marrow may be dormant or senescent and, as such, do not have clinical significance (50). In contrast to the aforementioned studies, we found that the presence of CEA mRNA-positive DTCs is a poor predictor for OS and CSS rates. The same correlation between DTCs and worse clinical outcome have shown in few studies [lower DFS (51), OS (52,53) and higher recurrence rate (51,54)].
Time of blood sampling
Although pulmonary vein ligation and interruption is the first step of surgery for NSCLC and the no-touch technique is used in all surgeries of malignant lung neoplasms, there is some evidence that tumor cells can disseminate during surgery (55-57). Hashimoto et al. have documented a significant CTC increase in pulmonary blood immediately after surgery. Using CellSearch, they found four CTCs in 2.5 mL of blood before surgery, which then increased up to 60 CTCs following surgery (56). However, the ability of CTCs to progress to metastasis remains unclear and blood sampling immediately after an operation can lead to erroneous conclusions. O’Sullivan et al. hypothesized that preoperative detection of CTCs reflects metastatic potential or residual disease (58). Sawabata et al. have shown the 2-year recurrence-free survival rates were 94.6% for patients without CTCs after surgery, 62.5% for patients with single CTCs, and 52.9% for patients with detected of CTCs clusters (P<0.01) (59). Therefore, in our study we evaluated the genuine (without iatrogenic effects) value of CTC/DTC status as an independent prognostic factor.
The correlation between CTCs and clinical-morphological criteria and the impact of CTCs on survival rate
In most previous studies, the presence of CTCs/DTCs is not correlated with the standard clinical and pathological characteristics of patients (44,46-49). However, Wang et al. (60) reported that CTCs were associated with locoregional lymph node metastases (OR =2.06; 95% CI: 1.18–3.62; P=0.027) and disease stage (OR=1.95; 95% CI: 1.08–3.54; P=0.011). We found no correlation between CTCs/DTCs and tumor size, lymph node metastasis, disease stage, histological type and malignancy level. We only found a connection between the presence of CTCs/DTCs and tobacco-smoking status, that was probably caused by low numbers of non-smokers in the cohort. Regardless of the utilized method and data, CTC presence use to be associated with a poor prognosis. Rossi et al. (61) demonstrated that CTC-positive patients, detected with an expanded CK panel, have significantly lower average OS (250 versus 767 days, respectively; P=0.033) and DFS (108 versus 254 days, respectively; P=0.017) than CTC-negative patients. Another study that used RT-PCR to identify CTCs based on the survivin marker showed that the presence of survivin-expressing CTCs is an independent predictor of cancer recurrence (HR =43.5; 95% CI: 2.67–70.9; P=0.008) and survival (HR =1.35; 95% CI: 1.02–4.31; P=0.049) (62). A recently conducted meta-analysis of 20 studies, which included 1,576 NSCLC patients, showed that CTCs can serve as a reliable prognostic marker for NSCLC. Despite the fact that the studies used different methods for detecting CTCs (such as RT-PCR, immunohistochemistry, CellSearch) the prognostic value of CTCs was nevertheless confirmed. A statistically confirmed connection between the presence of CTCs and decreased DFS (RR =2.14; 95% CI: 1.36–3.38; P<0.0001) and OS (RR =2.19; 95% CI: 1.53–3.12; P<0.0001) was found (60). The data from the presented study supports the previous published results.
Conclusions
In NSCLC patients undergoing radical surgery, we confirmed that the presence of CEA and EpCAM mRNA-positive CTCs/DTCs is associated with poorer survival (OS, CSS, DFS). Our study highlights the importance of CTCs/DTCs as a therapeutic target for adjuvant chemotherapy, biological and/or immunotherapy in early stage lung cancers.
Acknowledgments
Funding: This study was supported by Ministry of Health of the Czech Republic (No. NV18-03-00470), Ministry of Education, Youth and Sport of the Czech Republic (Nos. BBMRI – LM2018125, NCMG – LM2023067, EATRIS-CZ – LM2018133), Palacky University Olomouc (No. LF 2023_006), European Regional Development Fund (No. ACGT CZ.02.1.01/0.0/0.0/16_026/0008448), National Institute for Cancer Research (Programme EXCELES, ID Project No. LX22NPO5102) - Funded by the European Union - Next Generation EU and Cancer Research Czech Republic.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-801/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-801/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-801/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-801/coif). AR, MV, JSt, JD, JV, JSr and MH report that they obtained institutional funding by Ministry of Health of the Czech Republic (No. NV18-03-00470), Ministry of Education, Youth and Sport of the Czech Republic (Nos. BBMRI – LM2018125, NCMG - LM2023067, EATRIS-CZ – LM2018133), Palacky University Olomouc (No. LF 2023_006), European Regional Development Fund (No. ACGT CZ.02.1.01/0.0/0.0/16_026/0008448), National Institute for Cancer Research (Programme EXCELES, ID Project No. LX22NPO5102) - Funded by the European Union - Next Generation EU. JSr, RT and MH are co-founders of spin-off company Intellmed, Ltd., and Cancer Research Czech Republic Foundation. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics review board of University Hospital Olomouc and the Faculty of Medicine and Dentistry (IRB number 172/08) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012 v1.0 [En ligne]. [cité le 24 février 2020]. Disponible: https://publications.iarc.fr/Databases/Iarc-Cancerbases/GLOBOCAN-2012-Estimated-Cancer-Incidence-Mortality-And-Prevalence-Worldwide-In-2012-V1.0-2012
- Crosbie PA, Shah R, Krysiak P, et al. Circulating Tumor Cells Detected in the Tumor-Draining Pulmonary Vein Are Associated with Disease Recurrence after Surgical Resection of NSCLC. J Thorac Oncol 2016;11:1793-7. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. [Crossref] [PubMed]
- Skrzypski M, Dziadziuszko R, Szymanowska A, et al. The risk of distant metastases and prognosis prediction in early stage squamous cell lung cancer (SqCLC) by means of 3 microRNAs expression assessment. Eur Respir J 2011;38:1953.
- McWilliams A, Lam B, Sutedja T. Early proximal lung cancer diagnosis and treatment. Eur Respir J 2009;33:656-65. [Crossref] [PubMed]
- Pantel K, Cote RJ, Fodstad O. Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst 1999;91:1113-24. [Crossref] [PubMed]
- Franken B, de Groot MR, Mastboom WJ, et al. Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res 2012;14:R133. [Crossref] [PubMed]
- Iwao K, Watanabe T, Fujiwara Y, et al. Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small-cell lung cancer. Int J Cancer 2001;91:433-7. [Crossref] [PubMed]
- Matikas A, Syrigos KN, Agelaki S. Circulating Biomarkers in Non-Small-Cell Lung Cancer: Current Status and Future Challenges. Clin Lung Cancer 2016;17:507-16. [Crossref] [PubMed]
- Almufti R, Wilbaux M, Oza A, et al. A critical review of the analytical approaches for circulating tumor biomarker kinetics during treatment. Ann Oncol 2014;25:41-56. [Crossref] [PubMed]
- Ligthart ST, Coumans FA, Attard G, et al. Unbiased and automated identification of a circulating tumour cell definition that associates with overall survival. PLoS One 2011;6:e27419. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov 2016;6:479-91. [Crossref] [PubMed]
- Pathak R, Goldberg SB, Canavan M, et al. Association of Survival With Adjuvant Chemotherapy Among Patients With Early-Stage Non-Small Cell Lung Cancer With vs Without High-Risk Clinicopathologic Features. JAMA Oncol 2020;6:1741-50. [Crossref] [PubMed]
- Ghossein RA, Bhattacharya S, Rosai J. Molecular detection of micrometastases and circulating tumor cells in solid tumors. Clin Cancer Res 1999;5:1950-60. [PubMed]
- Banys-Paluchowski M, Krawczyk N, Meier-Stiegen F, et al. Circulating tumor cells in breast cancer--current status and perspectives. Crit Rev Oncol Hematol 2016;97:22-9. [Crossref] [PubMed]
- Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One 2014;9:e85350. [Crossref] [PubMed]
- Zhe X, Cher ML, Bonfil RD. Circulating tumor cells: finding the needle in the haystack. Am J Cancer Res 2011;1:740-51. [PubMed]
- Langley RR, Fidler IJ. The seed and soil hypothesis revisited--the role of tumor-stroma interactions in metastasis to different organs. Int J Cancer 2011;128:2527-35. [Crossref] [PubMed]
- Hosokawa M, Kenmotsu H, Koh Y, et al. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One 2013;8:e67466. [Crossref] [PubMed]
- Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol 2010;2010:617421. [Crossref] [PubMed]
- Cohen EN, Jayachandran G, Moore RG, et al. A Multi-Center Clinical Study to Harvest and Characterize Circulating Tumor Cells from Patients with Metastatic Breast Cancer Using the Parsortix(®) PC1 System. Cancers (Basel) 2022;14:5238. [Crossref] [PubMed]
- Lindsay CR, Blackhall FH, Carmel A, et al. EPAC-lung: pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur J Cancer 2019;117:60-8. [Crossref] [PubMed]
- Li J. Significance of Circulating Tumor Cells in Nonsmall-Cell Lung Cancer Patients: Prognosis, Chemotherapy Efficacy, and Survival. J Healthc Eng 2021;2021:2680526. [Crossref] [PubMed]
- Vidlarova M, Berta E, Prasil P, et al. Cannabinoid receptor 2 expression in early-stage non-small cell lung cancers identifies patients with good prognosis and longer survival. Transl Lung Cancer Res 2022;11:2040-50. [Crossref] [PubMed]
- Jancik S, Drabek J, Berkovcova J, et al. A comparison of Direct sequencing, Pyrosequencing, High resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas. J Exp Clin Cancer Res 2012;31:79. [Crossref] [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [Crossref] [PubMed]
- Koudelakova V, Kneblova M, Trojanec R, et al. Non-small cell lung cancer--genetic predictors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2013;157:125-36. [Crossref] [PubMed]
- Janku F, Srovnal J, Korinkova G, et al. Molecular detection of disseminated breast cancer cells in the bone marrow of early breast cancer patients using quantitative RT PCR for CEA. Neoplasma 2008;55:317-22. [PubMed]
- Havlik R, Srovnal J, Klos D, et al. Occult tumour cells in peritoneal lavage are a negative prognostic factor in pancreatic cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2013;157:233-8. [Crossref] [PubMed]
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009;55:611-22. [Crossref] [PubMed]
- Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Computational Statistics & Data Analysis 2003;43:121 37.
- Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compr Canc Netw 2010;8:740-801. [Crossref] [PubMed]
- Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Crosbie PA, Shah R, Summers Y, et al. Prognostic and predictive biomarkers in early stage NSCLC: CTCs and serum/plasma markers. Transl Lung Cancer Res 2013;2:382-97. [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel) 2014;6:153-65. [Crossref] [PubMed]
- Tamminga M, Andree KC, Hiltermann TJN, et al. Detection of Circulating Tumor Cells in the Diagnostic Leukapheresis Product of Non-Small-Cell Lung Cancer Patients Comparing CellSearch(®) and ISET. Cancers (Basel) 2020;12:896. [Crossref] [PubMed]
- Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. [Crossref] [PubMed]
- Okumura Y, Tanaka F, Yoneda K, et al. Circulating tumor cells in pulmonary venous blood of primary lung cancer patients. Ann Thorac Surg 2009;87:1669-75. [Crossref] [PubMed]
- Lv C, Zhao B, Wang L, et al. Detection of circulating tumor cells in pulmonary venous blood for resectable non-small cell lung cancer. Oncol Lett 2018;15:1103-12. [PubMed]
- Sienel W, Seen-Hibler R, Mutschler W, et al. Tumour cells in the tumour draining vein of patients with non-small cell lung cancer: detection rate and clinical significance. Eur J Cardiothorac Surg 2003;23:451-6. [Crossref] [PubMed]
- Sai B, Xiang J. Disseminated tumour cells in bone marrow are the source of cancer relapse after therapy. J Cell Mol Med 2018;22:5776-86. [Crossref] [PubMed]
- Brunsvig PF, Flatmark K, Aamdal S, et al. Bone marrow micrometastases in advanced stage non-small cell lung carcinoma patients. Lung Cancer 2008;61:170-6. [Crossref] [PubMed]
- Hsu CP, Shai SE, Hsia JY, et al. Clinical significance of bone marrow microinvolvement in nonsmall cell lung carcinoma. Cancer 2004;100:794-800. [Crossref] [PubMed]
- Poncelet AJ, Weynand B, Ferdin F, et al. Bone marrow micrometastasis might not be a short-term predictor of survival in early stages non-small cell lung carcinoma. Eur J Cardiothorac Surg 2001;20:481-8. [Crossref] [PubMed]
- Rud AK, Borgen E, Mælandsmo GM, et al. Clinical significance of disseminated tumour cells in non-small cell lung cancer. Br J Cancer 2013;109:1264-70. [Crossref] [PubMed]
- McGowan PM, Kirstein JM, Chambers AF. Micrometastatic disease and metastatic outgrowth: clinical issues and experimental approaches. Future Oncol 2009;5:1083-98. [Crossref] [PubMed]
- Cote RJ, Beattie EJ, Chaiwun B, et al. Detection of occult bone marrow micrometastases in patients with operable lung carcinoma. Ann Surg 1995;222:415-23; discussion 423-5. [Crossref] [PubMed]
- Sugio K, Kase S, Sakada T, et al. Micrometastasis in the bone marrow of patients with lung cancer associated with a reduced expression of E-cadherin and beta-catenin: risk assessment by immunohistochemistry. Surgery 2002;131:S226-31. [Crossref] [PubMed]
- Kasimir-Bauer S, Schleucher N, Weber R, et al. Evaluation of different markers in non-small cell lung cancer: prognostic value of clinical staging, tumour cell detection and tumour marker analysis for tumour progression and overall survival. Oncol Rep 2003;10:475-82. [Crossref] [PubMed]
- Sienel W, Mecklenburg I, Dango S, et al. Detection of MAGE-A transcripts in bone marrow is an independent prognostic factor in operable non-small-cell lung cancer. Clin Cancer Res 2007;13:3840-7. [Crossref] [PubMed]
- Sawabata N, Okumura M, Utsumi T, et al. Circulating tumor cells in peripheral blood caused by surgical manipulation of non-small-cell lung cancer: pilot study using an immunocytology method. Gen Thorac Cardiovasc Surg 2007;55:189-92. [Crossref] [PubMed]
- Hashimoto M, Tanaka F, Yoneda K, et al. Significant increase in circulating tumour cells in pulmonary venous blood during surgical manipulation in patients with primary lung cancer. Interact Cardiovasc Thorac Surg 2014;18:775-83. [Crossref] [PubMed]
- Sawabata N, Funaki S, Hyakutake T, et al. Perioperative circulating tumor cells in surgical patients with non-small cell lung cancer: does surgical manipulation dislodge cancer cells thus allowing them to pass into the peripheral blood? Surg Today 2016;46:1402-9. [Crossref] [PubMed]
- O'Sullivan GC, Collins JK, Kelly J, et al. Micrometastases: marker of metastatic potential or evidence of residual disease? Gut 1997;40:512-5. [Crossref] [PubMed]
- Sawabata N, Nakamura T, Kawaguchi T, et al. Circulating tumor cells detected only after surgery for non-small cell lung cancer: is it a predictor of recurrence? J Thorac Dis 2020;12:4623-32. [Crossref] [PubMed]
- Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One 2013;8:e78070. [Crossref] [PubMed]
- Rossi E, Tartarone A, Facchinetti A, et al. Non Small Cell Lung Cancer (NSCLC) and Circulating Tumor Cells (CTCs): Could an implemented CTC assay reveal higher risk patients? Ann Oncol 2015;26:vi80. [Crossref]
- Yie SM, Lou B, Ye SR, et al. Clinical significance of detecting survivin-expressing circulating cancer cells in patients with non-small cell lung cancer. Lung Cancer 2009;63:284-90. [Crossref] [PubMed]



