
Development of a cuproptosis-related signature for prognosis prediction in lung adenocarcinoma based on WGCNA
Highlight box
Key findings
• A novel cuproptosis-related gene signature was developed for the prognosis of lung adenocarcinoma (LUAD).
What is known and what is new?
• LUAD has a poor prognosis and improved therapies are needed. Cuproptosis is a newly discovered mitochondrial respiration-dependent cell death mechanism induced by copper that can kill cancer cells via copper carriers in cancer therapy.
• We successfully constructed a cuproptosis-related gene model to improve the prognostic evaluation of patients with LUAD.
What is the implication, and what should change now?
• This finding can provide a valid prognostic risk model for LUAD and improve understanding of its heterogeneity, which may assist in personalizing treatment and follow-up for patients.
Introduction
Non-small cell lung cancer (NSCLC) is one of the most common malignant tumors and can be divided into adenocarcinoma, squamous carcinoma, and large cell carcinoma (1). Lung adenocarcinoma (LUAD) is the most common lung cancer subtype and accounts for approximately 40% of lung cancer (2). According to statistics, LUAD has been the major cause of tumor-associated mortality in recent years (3). Previous studies have shown that patients with LUAD have a shorter survival time than do those with lung squamous cell carcinoma (LUSC) (4), as most cases of LUAD are found at the advanced stages (5). Despite new developments in cancer treatments in recent years, including surgical resection, immunotherapy, chemotherapy, and radiotherapy for LUAD (1), the prognosis of patients with LUAD continues to be disappointing, with the 5-year survival rate being less than 20% (6). Indeed, LUAD shows great inter- and intratumor heterogeneity, which is a clinical challenge driving tumor progression and drug resistance (7). Therefore, there is a need to explore novel prognostic markers for guiding the genetic classification and clinical treatment of LUAD.
Copper is an essential mineral nutrient for living organisms, but dysregulation of copper stores, either overload or deficiency, can induce oxidative stress and cytotoxicity (8). Diverse functions of mitochondria have been revealed, such as producing biosynthetic intermediates and contributing to cellular stress responses (9). Since the assembly of cuproenzymes in mitochondria requires copper, this mineral plays a crucial role in mitochondrial function and signaling, which can induce multiple forms of cell death, including apoptosis and autophagy (10). Recently, researchers from the Broad Institute of Harvard and MIT presented a copper-induced cell death pathway, named cuproptosis, which is mediated by protein lipoylation, and several genes involved in cuproptosis were identified. Copper binds to the lipoacylated components of the tricarboxylic acid (TCA) cycle, which causes abnormal aggregation of lipoylated proteins and loss of Fe-S cluster–containing proteins, leading to proteotoxic stress and ultimately cell death (11). A study has demonstrated the association of copper imbalances with tumor burden, progression, incidence, invasion, and reoccurrence of the disease (10). A previous study revealed a long non-coding RNA signature related to cuproptosis and could predict clinical outcomes of LUAD (12). But the effect of serum copper levels on lung cancer risk remains controversial (13,14). Moreover, the impact of cuproptosis-related genes on cancer progression and the clinical outcome in LUAD is still poorly understood.
Weighted gene co-expression network analysis (WGCNA) is a systems biology method that can describe the correlation of gene expression patterns across different samples (15). WGCNA is based on two hypotheses, the first being that genes with similar expression patterns may be coregulated, functionally related, or located in the same pathway; and the second being that the gene network fits into a scale-free distribution. In many studies, WGCNA has been used as a powerful method to explore the complex relationships between gene expression profiles and phenotypes (16). It can be used to find clusters of highly correlated genes and to summarize clusters or associate different modules using the module eigengene (15). Frequently, WGCNA is used in examining various biological processes, which is quite helpful for the identification of candidate biomarkers or therapeutic targets (17). A recent study investigated prognosis-associated key genes in LUAD based on WGCNA (18), which inspired us to try to excavate the cuproptosis-related prognostic genes.
In this study, we calculated cuproptosis-related gene set enrichment scores for tumor samples using single-sample gene-set enrichment analysis (ssGSEA) based on The Cancer Genome Atlas (TCGA)-LUAD data set. Modules associated with cuproptosis were identified by WGCNA and correlation analysis. Univariate Cox regression analysis was used to identify prognosis-related genes, the least absolute shrinkage and selection operator (LASSO) Cox regression model was used to construct a prognostic risk model for LUAD, and validation was completed with the GSE72094 data set. The median prognostic risk score was used to stratify patients, with overall survival (OS), tumor characteristics, and immune cell infiltration being compared between the high-risk and low-risk groups. Furthermore, correlation analysis of prognostic risk score and half-maximum inhibitory concentration (IC50) values of drugs in The Genomics of Drug Sensitivity in Cancer (GDSC) v. 2 database revealed different drug sensitivities across the different risk groups. Collectively, our findings could help researchers to better understand the function of cuproptosis in the development of LUAD and may inform the prognostic prediction and personalized treatment of LUAD. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-157/rc).
Methods
Data collection
The cuproptosis gene set was obtained from the study of Tsvetkov et al. (11) and included FDX1, LIAS, LIPT1, DLD, DLAT, PDHA1, PDHB, MTF1, GLS, and CDKN2A. RNA sequencing, copy number aberration (CNA), and single-nucleotide variation (SNV) data from TCGA database (https://portal.gdc.cancer.gov; accessed December 2021) of LUAD tumors, and microarray data GSE72094 from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; accessed December 2021) were downloaded. In addition, the standardized clinical information was extracted from TCGA Pan-Cancer Clinical Data Resource data set (19). A total of 497 TCGA-LUAD tumor samples with both sequencing data and survival information were retained for subsequent analysis, and the sample statistics are shown in Table 1. The GEO data set was processed as follows: (I) probes were matched to the gene symbols using the annotation provided by the platform; (II) probes corresponding to multiple genes were removed; (III) if multiple probes matched the same gene symbol, the median was considered as the expression value. Transcription profiles from non-small-cell lung cancer patients during anti-PD-1 treatment were also obtained from the GEO database (GSE126044). According to the RECIST 1.1 criteria, patients were evaluated and classified: as responders and non-responders: patients who showed a partial response (PR) or stable condition (SC) over 6 months were classified as responders, and patients who show progressive disease (PD) or SC within 6 months or less are classified as non-responders.
Table 1
| Characteristics | Variables | Number of samples (N=497) |
|---|---|---|
| Age | <60 years | 146 (29.4%) |
| ≥60 years | 351 (70.6%) | |
| ALK_eml4 | No | 206 (41.4%) |
| Yes | 33 (6.6%) | |
| Unknown | 258 (51.9%) | |
| EGFR | Mut | 79 (15.9%) |
| WT | 190 (38.2%) | |
| Unknown | 228 (45.9%) | |
| Gender | Female | 269 (54.1%) |
| Male | 228 (45.9%) | |
| Stage | I/II | 385 (77.5%) |
| III/IV | 105 (21.1%) | |
| Unknown | 7 (1.4%) | |
| Smoking | Yes | 408 (82.1%) |
| No | 71 (14.3%) | |
| Unknown | 18 (3.6%) |
ALK and EGFR are genes that are frequently mutated in lung cancer. TCGA, The Cancer Genome Atlas; Mut, mutation; WT, wild type.
Identification of cuproptosis-related hub genes
ssGSEA was performed to derive the enrichment score of the cuproptosis gene set using the R package “GSVA” (The R Foundation for Statistical Computing), and the cuproptosis Z-score of each sample in the TCGA-LUAD cohort was calculated. Then, the WGCNA was performed with the “WGCNA” R package to build the co-expression modules and clarify the relationship of the underlying modules. To meet the scale-free topology criteria for optimal clustering, the soft-thresholding power beta was chosen with a scale-free fit index of 0.85. Modules were detected with hierarchical clustering analysis and the mixed dynamic cut tree algorithm, and the minimum number of genes for each module was set to 30. Pearson correlation analysis was used to find those modules significantly associated with cuproptosis Z-score. Finally, cuproptosis-related hub genes with a module membership (MM) >0.6 and a gene significance (GS) >0.2 were selected [MM measures the relationship of a gene with the particular module, and GS incorporates external information into the co-expression network (15)]. Finally, to explore the functional characteristics of the hub genes, biological function enrichment analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was applied using the “clusterProfiler” R package. When the P value was less than 0.05, the enriched pathway was considered to be statistically significant.
Construction of the cuproptosis-related prognostic model
Univariate Cox regression analysis was performed on the hub genes using the R package “survival” to identify genes related to prognosis (P<0.01), and forest plots were created using the package “forestplot”. Then, the R package “glmnet” was used for LASSO Cox regression analysis to further screen the key prognosis-related genes. The LASSO algorithm yields a reliable model by shrinking and selecting the variables, making some regression coefficients strictly equal to 0, thus preventing overfitting. At last, we established a prognostic model and calculated the risk scores of the patients according to the normalized expression level of each key prognosis-related gene and its corresponding regression coefficients. The formula was established as follows:
where Exp (i) represents the expression level of each gene, and Coef (i) indicates the corresponding regression coefficient.
Patients were divided into high-risk and low-risk score groups according to the median value. Survival analysis was performed using the R package “survminer”, and receive operating characteristic (ROC) curves were plotted with the R package “timeROC” to assess the efficacy of the prognostic model. Furthermore, we performed univariate and multivariate Cox analyses to explore the independent prognostic value of the risk score.
Analysis of immune cell infiltration in the tumor microenvironment (TME)
Signature genes of 28 immune cell types including activated CD8 T cells, activated dendritic cells, and macrophages were acquired from a previous study (20). We used the ssGSEA algorithm in the R package “GSVA” (21) to assess the proportions of 28 types of infiltrating immune cells based on TCGA-LUAD gene expression RNA-sequencing (RNA-seq) data. The ssGSEA algorithm is a rank-based method that defines a score representing the degree of absolute enrichment of a particular gene set in each sample (22). In addition, the ssGSEA scores for immune cells in different groups were also compared using the Wilcoxon rank-sum test. Furthermore, we applied the Estimation of Stromal and Immune cells in Malignant Tumors using Expression data (ESTIMATE) algorithm in the R package “estimate” to assess the immune scores, stromal scores, ESTIMATE scores, and tumor purity for each LUAD sample (23).
Gene set variation analysis
To explore differences in cell signaling pathways between high and low risk groups, the gene sets h.all.v7.4.symbols.gmt and c2.cp.kegg.v7.4.symbols.gmt were downloaded from the GSEA website (https://www.gsea-msigdb.org/gsea/downloads.jsp). |normalized enrichment score (NES)|>1, P<0.05, and FDR q<0.25 were considered significant. We use the “pheatmap” R package to display the results.
Drug sensitivity prediction
The GDSC (http://www.cancerrxgene.org; accessed March 2022) database is the largest public resource for information on drug sensitivity in cancer cells and molecular markers of drug response (24). To explore the treatment response in different risk groups, drug response prediction was conducted in R by using the “oncoPredict” package based on the cell line expression data and response information in the GDSC v. 2 database and the RNA-seq data of the TCGA-LUAD data set. IC50 indicates the effectiveness of a substance in inhibiting specific biological or biochemical functions. In this study, we estimated the IC50 of drugs in the GDSC v. 2 database for each patient with LUAD and calculated the Spearman rank correlation coefficient between IC50 values and prognostic risk scores.
Statistical analysis
All statistical analyses were implemented using R version 4.1.2. The Wilcoxon rank-sum test was used to identify differentially expressed genes and filter infiltrating immune cells. Univariable Cox and LASSO Cox regression analyses were used to construct a prognostic risk model. The Kaplan-Meier method was used to generate survival curves for the high-risk and the low-risk groups, and the log-rank test was used to determine statistically significant differences. To test the predictive ability of the prognostic signature, the area under the ROC curve (AUC) was calculated. For all analyses, P values lower than 0.05 were considered statistically significant.
Ethical statement
The relevant data provided by TCGA and GEO databases are publicly available and open-ended, and do not require the approval of the local ethics committee. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Mutational landscape of the cuproptosis-related gene set
To gain a fundamental understanding of the cuproptosis process in tumor development, we performed somatic mutation and CNV analysis to detect the alterations of 10 cuproptosis-related genes in 527 TCGA-LUAD samples. Among the 527 samples, 42 (7.97%) harbored somatic mutations of the cuproptosis gene set. The mutation frequency of CDKN2A (3%) was the highest, followed by DLAT and DLD (1%). Missense mutation was the most common mutation type, and splice mutation occurred in CDKN2A and PDHA1 (Figure 1A). CNV events were general in the cuproptosis gene set, and amplifications were more frequent in MTF1, GLS, and LIAS, while deletions were more frequent in CDKN2A, FDX1, and DLAT (Figure 1B). Notably, the prevalence of CNV loss in CDKN2A exceeded 20%. CDKN2A was a cuproptosis suppressor gene, and homozygous deletion of CDKN2A was associated with shorter disease-free survival and OS (25).

We explored the association between the expression of the cuproptosis gene set and multiple clinical factors, including age, gender, tumor stage, and status. As shown in Figure S1, the expression of DLAT and LIPT1 were significantly different in the age ≥60 years group and the age <60 years group; the expression levels of DLAT, PHDB, and DLD were higher in male patients than in females, while GLS and LIPT1 showed the opposite tendency. DLAT, DLD, and PDHB were highly expressed in tumor stage III–IV compared with stage I–II. All of the 10 genes were differentially expressed in the tumor group versus the normal group.
Identification of hub genes and functional enrichment analysis
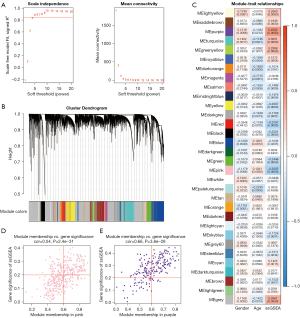
First, we calculated the cuproptosis Z-score for each patient in the TCGA-LUAD cohort using ssGSEA. Then, co-expression modules were built with WGCNA, and Pearson correlation was applied to evaluate the relationships of the underlying modules, cuproptosis Z-score, and clinical traits (age, gender). As a result, we identified 32 modules with the optimal soft threshold power =5 (Figure 2A,2B). Of the 32 modules, 21 were significantly correlated with cuproptosis Z-scores (P<0.05), of which 8 were positively correlated and 13 were negatively correlated (Figure 2C). To screen for the hub genes associated with cuproptosis, we selected the MEpurple module with the most significant positive correlation (Pearson correlation coefficient = 0.458) and the MEpink module with the most significant negative correlation (Pearson correlation coefficient = –0.326). A total of 238 hub genes with MM >0.6 and GS >0.2 were obtained, including 107 genes in the MEpurple module and 131 genes in the MEpink module (Figure 2D,2E).
Subsequently, biological function enrichment analysis including GO and KEGG pathway enrichment analyses were conducted to explore the functional characteristics of the 238 hub genes (Figure 3). The GO analysis results revealed that the hub genes were significantly enriched in the mitochondrial inner membrane term, and mitochondrial protein-containing complex term related to cell component (CC), which corresponded with the fact that copper-induced cell death was dependent on mitochondrial respiration. Molecular function (MF) terms of the hub genes included extracellular matrix structural constituent, cytokine binding, and transforming growth factor (TGF) beta binding, while biological process (BP) terms mainly included endothelium development, extracellular matrix assembly, smooth muscle cell differentiation, and lymph vessel development. According to previous reports, TGF-beta is an important regulator of the extracellular matrix in the developing lung (26), and cells and extracellular matrix in the airway responded both passively and actively to the mechanical stimulation induced by smooth muscle contraction in chronic obstructive pulmonary disease (27). KEGG signaling pathways included mainly cell adhesion molecules, cGMP−PKG signaling pathway, oxidative phosphorylation, and diabetic cardiomyopathy. Cell adhesion molecules have been shown to play an important role in inflammatory response, angiogenesis, and tumor progression (28).

Construction of the prognostic risk model for LUAD
The prognostic effects of 238 hub genes were analyzed using univariate Cox regression, and 35 prognosis-related hub genes were screened out for subsequent model construction (P<0.01). The TCGA-LUAD patients were divided into high-risk and low-expression groups according to the median expression values of each prognosis-related gene. The forest plot and corresponding survival curves of the top 10 genes are shown in Figure S2. LASSO Cox regression was then performed to reduce the overfit and the number of genes for further analysis (Figure S3A). Afterward, we screened out the 7 best predictors associated with LUAD prognosis, which included PTGES3, NMUR1, CLEC3B, METTL7A, DNAAF9, C1QTNF7, and RPE, with PTGES3 and RPE being the risk factors and the others being the protective factors (Figure S3B). The two risk genes belong to the MEpurple module that showed the most significant positive correlation with cuproptosis. The detailed functional annotation analysis (website: https://cdn.amegroups.cn/static/public/tlcr-23-157-01.xlsx) indicated the two risk genes tend to act in the intracellular anatomical structure and are involved in cellular metabolic processes which might further regulate the cuproptosis process. On the contrary, the five protective genes belong to the MEpink module with the most significant negative correlation with cuproptosis. And the five protective genes are involved in response to stimulus and regulation of BP, they tend to function in the cell periphery and extracellular matrix. Expression levels of these genes and corresponding coefficients derived from the LASSO Cox regression model were used to calculate the risk score for each patient as following: Risk Score = RPE × 0.1371 – CLEC3B × 0.1338 – METTL7A × 0.0873 + PTGES3 × 0.2231 – NMUR1 × 0.2774 – C1QTNF7 × 0.007 – DNAAF9 × 0.0162. The TCGA-LUAD patients were divided into high-risk and low-risk groups based on the median risk score. Kaplan-Meier analysis indicated that patients in the low-risk group experienced significantly better survival than did those in the high-risk group (Figure 4A). AUC of the risk model was 0.682 at 1 year, 0.637 at 3 years, and 0.692 at 5 years (Figure 4B). In addition, the 7 genes were differentially expressed between the high-risk and low-risk groups, PTGES3 and RPE were highly expressed in the high-risk group, and the other 5 genes were highly expressed in the low-risk group (Figure 4C-4E).

External validation of the prognostic risk model
To validate the predictive value and robustness of the prognostic risk model, we downloaded the GSE72094 data set as a validation set and calculated the risk score with the same formula for each patient with LUAD. Samples in GSE72094 were divided into high-risk and low-risk groups according to the median risk score, and consistent with the results in the TCGA cohort, patients in the high-risk group showed significantly poorer OS than did the patients in the low-risk group (Figure S4A). The AUCs for 1-year, 3-year, and 5-year OS were 0.641, 0.665, and 0.719, respectively (Figure S4B). Prognostic risk genes were also highly expressed in the high-risk group, while protective genes were highly expressed in the low-risk group (Figure S4C-S4E). Overall, the results demonstrated that the risk score could predict OS in LUAD.
Independent prognostic role of the risk score
To explore the independent prognostic value of the risk score and several clinicopathological variables (gender, age, TNM stage and smoking), univariate and multivariate Cox regression analyses were performed on the TCGA-LUAD data set. The results indicated that the TNM stage and risk score calculated from the prognostic risk model were independent prognostic factors for OS (Figure S5A). The risk score was consistently an independent prognostic factor in the GSE72094 cohort (Figure S5B). Furthermore, we completed the Wilcoxon rank-sum test and Kruskal-Wallis test to investigate whether our risk model correlated with the clinical characteristics and found that the high-risk group had a more advanced TNM stage and a higher proportion of males (Figure S5C-S5J).
Tumor immunity relevance analysis
Gene set variation analysis (GSVA) was performed based on the hallmark gene sets obtained from the MsigDB database (https://www.gsea-msigdb.org/gsea/index.jsp) to explore the molecular mechanisms of different risk groups in the TCGA-LUAD cohort. We found that the enrichment scores of most gene sets including ADIPOGENESIS, ANDROGEN RESPONSE, COAGULATION, MTORC1 SIGNALING, P53 PATHWAY, and PI3K AKT MTOR SIGNALING varied significantly between high-risk and low-risk groups (Figure S6). A previous study has revealed that the accumulation of copper inhibits the PI3K/AKT/mTOR pathway, which activates cellular autophagy and disrupts mitochondrial dynamics (29). Moreover, a high dose of copper can activate p53-independent apoptosis through the induction of nucleolar stress in human cell lines (30), yet the role of the p53 pathway in cuproptosis remains understudied.
Further, the association between the risk score and immune response was investigated. At first, we obtained the ESTIMATE score, immune score, stromal score, and tumor purity of each TCGA-LUAD patient based on the ESTIMATE algorithm (23). The high-risk group showed significantly lower ESTIMATE score, immune score, and stromal score, which indicated fewer immune and stroma cell infiltrated. In contrast, tumor purity was significantly higher than in the low-risk group (Figure 5A). Spearman correlation analysis supported the risk score negatively associated with immune and stromal cell infiltration while positively associated with tumor purity (Figure 5B). Then we calculated infiltration level of specific immune cell types based on the expression of their signature genes using ssGSEA. We found most of the immune cell types were reduced in the high-risk group. However, it enriched infiltration of CD56 bright NK cells, activated CD4 T cells, effector memory CD4 T cells, and memory B cells (Figure 5C). It is known that the cytotoxic activity of CD56 bright NK cells is significantly lower than that of CD56 dim cells and they express less perforin, granzymes and cytolytic granules (31). These results suggested that a suppressed or not fully activated immune microenvironment in the high-risk group with highly expressed cuproptosis indicator genes.

In addition, we investigated the potential of the risk score for predicting immunotherapy response. We obtained the transcription profiles from 18 non-small-cell lung cancer patients during anti-PD-1 treatment from the GEO database (GSE126044) and compared the risk score between responders and non-responders. The results indicated a significantly higher risk score in non-responders (Wilcoxon rank-sum test P=0.038). Furthermore, the predictive efficiency of the risk score for immunotherapy response achieved AUC 0.801 (Figure S7). These results suggested the cuproptosis-related risk score could further serve as a potential biomarker for immunotherapy response.
Besides, we analyzed the genomic and transcriptomic variations including (SNVs and CNVs in the high-risk and low-risk groups. The occurrence of CNV in TCGA-LUAD is summarized in Figure 6A and Figure 6B. The analysis of CNV alteration indicated a higher frequency in the high-risk group (Figure 6C). An oncoplot showed the most frequently mutated genes in the high-risk and low-risk groups (Figure 6D). As illustrated, TP53 (48%), TTN (45%), MUC16 (39%), CSMD3 (37%), and RYR2 (36%) were more frequently mutated and presented at a higher mutation frequency in the high-risk group.

Drug sensitivity analysis
Due to the high heterogeneity of LUAD, different patients have different therapeutic responses to the same drug. In this study, we predicted the sensitivity of TCGA-LUAD patients to drugs in the GDSC v. 2 database using the R package “oncoPredict” based on the IC50 of each sample. The GDSC v. 2 database is the largest public resource for information on drug sensitivity in cancer cells and molecular markers of drug response, and contains information on the sensitivity and response of different tumor cells from the cell, drug, and molecule levels. Results of the Spearman correlation analysis between IC50 and the risk score showed that the estimated IC50s differed significantly between the 2 groups, suggesting a greater sensitivity to PRT062607, BMS-754807, doramapimod, GSK269962A, ribociclib, and nutlin-3a (−) when the low-risk group had lower IC50s than those of the high-risk group (Figure 7A); meanwhile, BI-2536, UMI-77, MK-1775, savolitinib, docetaxel, and sepantronium were more effective for high-risk patients (Figure 7B).

Discussion
LUAD is one of the most aggressive and rapidly lethal tumor types, and patients with this disease have an OS of less than 5 years (32). Over the past few decades, many newly identified biomarkers or gene signatures have been found that have the potential to predict the prognosis of LUAD, and novel treatments for LUAD, including targeting therapy, immunotherapy, chemotherapy, and radiotherapy, have been developed (6). However, the prognosis of LUAD continues to be poor for the following reasons: (I) the effect of these strategies is limited by inadequate knowledge of the biological features of lung cancer, (II) most prognostic signatures are still in the molecular research phase and have not yet been applied in clinical practice, (III) treatment response varies widely between patients due to the heterogeneity of LUAD, and (IV) a large proportion of patients with LUAD are found at an advanced stage (1,5,6). Thus, uncovering reliable prognostic indicators for the prognosis of LUAD would be of great clinical significance. Cuproptosis is a newly recognized type of programmed cell death, in which intracellular copper accumulation triggers the aggregation of mitochondrial lipoylated proteins and the destabilization of the Fe–S cluster proteins, thus leading to cell death (11). A growing body of evidence has linked copper signaling to cell proliferation, tumor growth, and tumor metastasis. For example, elevated serum copper levels may increase the risk of lung cancer, as serum copper levels were found to be higher in patients in LUAD than in controls (8,10). Although many studies have revealed the association between copper and cancer development, the role of cuproptosis in LUAD has not yet been elucidated. Therefore, it is essential to identify robust signatures to enhance the prognostic prediction of patients with LUAD. This work thus established a novel cuproptosis-related 7-gene prognostic signature in the TCGA-LUAD cohort and validated it in the GSE72094 data set.
In the study, we obtained 10 cuproptosis genes identified in a previous study (11), and ssGSEA was used to calculate the cuproptosis Z-score for each LUAD patient in the TCGA database. Meanwhile, 32 co-expression modules were built by WGCNA, and the Pearson correlation coefficient was applied to evaluate the relationships between these modules and the cuproptosis Z-score. Subsequently, the MEpurple module and MEpink module were selected as being most significantly correlated with the Z-score. Functional enrichment analysis of 238 hub genes in the 2 modules showed that they were involved in a series of cuproptosis and LUAD pathogenesis-related pathways, such as TGF-beta binding, cell adhesion molecules, and mitochondrial inner membrane. Therefore, focusing on these biological functions and pathways would help us elucidate the underlying mechanisms of cuproptosis in LUAD. Next, we analyzed the prognostic values of 238 hub genes in TCGA data set using univariate Cox regression, and 35 hub genes were found to be significantly correlated with OS. Furthermore, the 7 genes most associated with prognosis were identified by LASSO Cox regression and included PTGES3, NMUR1, CLEC3B, METTL7A, DNAAF9, C1QTNF7, and RPE. Moreover, high expression of NMUR1, CLEC3B, METTL7A, DNAAF9, and C1QTNF7 were found to be associated with good prognosis, while high expression of PTGES3 and RPE were found to be associated with poor prognosis. We constructed a novel risk signature based on the 7 genes. CLEC3B is a member of the CLEC family, and abnormal expression of CLEC3B is related to many cancer types, with a lower expression of CLEC3B in LUAD being associated with attenuated cell adhesion capacity (33). Furthermore, the protein encoded by CLEC3B may be related to calcium ion binding which can affect copper binding (34). Gao et al. found PTGES3 messenger RNA (mRNA) and protein expression to be significantly elevated in LUAD tissues compared with normal lung tissues (35). Ma et al. revealed that the downregulated NMUR1 in patients with LUAD negatively affected the OS rate and that the high expression of NMUR1 might facilitate antitumor immunity in the LUAD microenvironment (36). Guo et al. reported METTL7A to be significantly downregulated and involved in the development of LUAD (37). Furthermore, the expression and the prognostic role of the 7 genes at the protein level warrant further investigation. Our results, viewed in conjunction with the literature, suggest that the prognostic signature is linked with LUAD. Low-risk scores and high-risk scores show significant survival differences, and patients with high-risk scores have worse outcomes in LUAD.
The TCGA-LUAD samples were divided into high-risk and low-risk groups according to the median risk score, and patients in the high-risk group showed significantly poorer survival than did the patients in the low-risk group. In addition, the 7 genes were found to be differentially expressed between the 2 groups, and prognostic risk genes PTGES3 and RPE were highly expressed in the high-risk group while the other 5 protective genes were highly expressed in the low-risk group. Additionally, the prognosis predictive and independent performance of our signature was validated not only in the TCGA-LUAD cohort but also in the GSE72094 cohort. Therefore, our prognostic risk model might be an effective marker for LUAD prognosis prediction.
The TME has garnered increased attention due to its association with tumor growth, invasion, and metastasis (38). Furthermore, different immune cell types play different roles in the antitumor and tumor immune escape processes (39). In the present study, ssGSEA was employed to investigate the relationship between our prognostic model and immune cell infiltration. Higher infiltration levels of activated CD4 T cells, activated dendritic cells, and memory B cells were observed in the high-risk patients. The high proportion of eosinophil, natural killer cells, and mast cells displayed a negative correlation with the prognostic model, revealing that the model may serve as a predictor for increased immune cell infiltration. Furthermore, the result of the ESTIMATE analysis showed that the high-risk group had lower immune scores and higher tumor purity. We also detected significant differences in the mutation profiles between the high- and low-risk groups: the high-risk group of LUAD tended to have a higher frequency of CNVs and SNVs, especially in TP53, TTN, and MUC16. TP53 encodes a tumor-suppressor protein, and mutations in this gene are associated with a variety of human cancers. Mutation of TP53 occurs in approximately 50.00% of patients with LUAD and leads to a weakened immune response in early-stage LUAD (39). Wu et al. demonstrated that patients with LUAD and MUC16 peptide mutants have worse outcomes (40). TTN encodes a large abundant protein of striated muscle, and the expression of TTN has been found to be significantly lower in LUAD and to be associated with worse survival (41). Additionally, the TTN mutant has been linked to high immunogenicity and inflammation of the TME in patients with LUAD (42).
Finally, drug prediction was completed using the R package “oncoPredict” to identify different candidate drugs for high-risk and low-risk patients. Drugs including PRT062607, BMS-754807, doramapimod, GSK269962A, ribociclib, and nutlin-3a (−) appeared to be more effective for the low-risk group, while the high-risk group appeared to be more sensitive to BI-2536, UMI-77, MK-1775, savolitinib, docetaxel, and sepantronium. As personalized therapy emerges into the forefront of cancer treatment, our study provides a potential tool for the selection of therapeutic approaches for patients with LUAD.
Conclusions
We used comprehensive bioinformatics analysis to develop a prognostic signature based on cuproptosis-related genes, thus providing a potential tool to predict the clinical prognosis of patients with LUAD.
Acknowledgments
Funding: This study was supported by National Natural Science Foundation of China (No. 82172786) and the Haiyan Foundation of Harbin Medical University Cancer Hospital (No. JJMS2023-04).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-157/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-157/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-157/coif).
Ethical Statement: The authors are accountable for all aspects of the works in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved. The relevant data provided by TCGA and GEO database are publicly available and open-ended, and do not require the approval of the local ethics committee. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang H, Guo L, Chen J. Rationale for Lung Adenocarcinoma Prevention and Drug Development Based on Molecular Biology During Carcinogenesis. Onco Targets Ther 2020;13:3085-91. [Crossref] [PubMed]
- Yi M, Li A, Zhou L, et al. Immune signature-based risk stratification and prediction of immune checkpoint inhibitor's efficacy for lung adenocarcinoma. Cancer Immunol Immunother 2021;70:1705-19. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Sun GZ, Zhao TW. Lung adenocarcinoma pathology stages related gene identification. Math Biosci Eng 2019;17:737-46. [Crossref] [PubMed]
- Inamura K. Clinicopathological Characteristics and Mutations Driving Development of Early Lung Adenocarcinoma: Tumor Initiation and Progression. Int J Mol Sci 2018;19:1259. [Crossref] [PubMed]
- Song J, Sun Y, Cao H, et al. A novel pyroptosis-related lncRNA signature for prognostic prediction in patients with lung adenocarcinoma. Bioengineered 2021;12:5932-49. [Crossref] [PubMed]
- Seguin L, Durandy M, Feral CC. Lung Adenocarcinoma Tumor Origin: A Guide for Personalized Medicine. Cancers (Basel) 2022;14:1759. [Crossref] [PubMed]
- Ge EJ, Bush AI, Casini A, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer 2022;22:102-13. [Crossref] [PubMed]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012;148:1145-59. [Crossref] [PubMed]
- Ruiz LM, Libedinsky A, Elorza AA. Role of Copper on Mitochondrial Function and Metabolism. Front Mol Biosci 2021;8:711227. [Crossref] [PubMed]
- Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375:1254-61. [Crossref] [PubMed]
- Ma S, Zhu J, Wang M, et al. A cuproptosis-related long non-coding RNA signature to predict the prognosis and immune microenvironment characterization for lung adenocarcinoma. Transl Lung Cancer Res 2022;11:2079-93. [Crossref] [PubMed]
- Zhang X, Yang Q. Association between serum copper levels and lung cancer risk: A meta-analysis. J Int Med Res 2018;46:4863-73. [Crossref] [PubMed]
- Jin Y, Zhang C, Xu H, et al. Combined effects of serum trace metals and polymorphisms of CYP1A1 or GSTM1 on non-small cell lung cancer: a hospital based case-control study in China. Cancer Epidemiol 2011;35:182-7. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Tan R, Zhang G, Liu R, et al. Identification of Early Diagnostic and Prognostic Biomarkers via WGCNA in Stomach Adenocarcinoma. Front Oncol 2021;11:636461. [Crossref] [PubMed]
- Wan Q, Tang J, Han Y, et al. Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp Eye Res 2018;166:13-20. [Crossref] [PubMed]
- Zhu H, Yue H, Xie Y, et al. Bioinformatics and integrated analyses of prognosis-associated key genes in lung adenocarcinoma. J Thorac Dis 2021;13:1172-86. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-416.e11. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Zuo S, Wei M, Wang S, et al. Pan-Cancer Analysis of Immune Cell Infiltration Identifies a Prognostic Immune-Cell Characteristic Score (ICCS) in Lung Adenocarcinoma. Front Immunol 2020;11:1218. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Peng Y, Chen Y, Song M, et al. Co-occurrence of CDKN2A/B and IFN-I homozygous deletions correlates with an immunosuppressive phenotype and poor prognosis in lung adenocarcinoma. Mol Oncol 2022;16:1746-60. [Crossref] [PubMed]
- Deng S, Zhang H, Han W, et al. Transforming Growth Factor-β-Neutralizing Antibodies Improve Alveolarization in the Oxygen-Exposed Newborn Mouse Lung. J Interferon Cytokine Res 2019;39:106-16. [Crossref] [PubMed]
- Bidan CM, Veldsink AC, Meurs H, et al. Airway and Extracellular Matrix Mechanics in COPD. Front Physiol 2015;6:346. [Crossref] [PubMed]
- Jang JH, Yang ES, Min KJ, et al. Inhibitory effect of butein on tumor necrosis factor-α-induced expression of cell adhesion molecules in human lung epithelial cells via inhibition of reactive oxygen species generation, NF-κB activation and Akt phosphorylation. Int J Mol Med 2012;30:1357-64. [Crossref] [PubMed]
- Wang Y, Zhao H, Shao Y, et al. Copper or/and arsenic induces autophagy by oxidative stress-related PI3K/AKT/mTOR pathways and cascaded mitochondrial fission in chicken skeletal muscle. J Inorg Biochem 2018;188:1-8. [Crossref] [PubMed]
- Chen CH, Chou YT, Yang YW, et al. High-dose copper activates p53-independent apoptosis through the induction of nucleolar stress in human cell lines. Apoptosis 2021;26:612-27. [Crossref] [PubMed]
- Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol 2001;22:633-40. [Crossref] [PubMed]
- Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 2018;9:117. [Crossref] [PubMed]
- Lu X, Shen J, Huang S, et al. Down-regulation of CLEC3B facilitates epithelial-mesenchymal transition, migration and invasion of lung adenocarcinoma cells. Tissue Cell 2022;76:101802. [Crossref] [PubMed]
- Iglesias A, López R, Fiol S, et al. Analysis of copper and calcium-fulvic acid complexation and competition effects. Water Res 2003;37:3749-55. [Crossref] [PubMed]
- Gao P, Zou K, Xiao L, et al. High expression of PTGES3 is an independent predictive poor prognostic biomarker and correlates with immune infiltrates in lung adenocarcinoma. Int Immunopharmacol 2022;110:108954. [Crossref] [PubMed]
- Ma Y, Zou H. Identification of the circRNA-miRNA-mRNA Prognostic Regulatory Network in Lung Adenocarcinoma. Genes (Basel) 2022;13:885. [Crossref] [PubMed]
- Guo T, Ma H, Zhou Y. Bioinformatics analysis of microarray data to identify the candidate biomarkers of lung adenocarcinoma. PeerJ 2019;7:e7313. [Crossref] [PubMed]
- Mao X, Xu J, Wang W, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 2021;20:131. [Crossref] [PubMed]
- Wang L, He T, Liu J, et al. Revealing the Immune Infiltration Landscape and Identifying Diagnostic Biomarkers for Lumbar Disc Herniation. Front Immunol 2021;12:666355. [Crossref] [PubMed]
- Wu C, Rao X, Lin W. Immune landscape and a promising immune prognostic model associated with TP53 in early-stage lung adenocarcinoma. Cancer Med 2021;10:806-23. [Crossref] [PubMed]
- Chen J, Wen Y, Su H, et al. Deciphering Prognostic Value of TTN and Its Correlation With Immune Infiltration in Lung Adenocarcinoma. Front Oncol 2022;12:877878. [Crossref] [PubMed]
- Wang Z, Wang C, Lin S, et al. Effect of TTN Mutations on Immune Microenvironment and Efficacy of Immunotherapy in Lung Adenocarcinoma Patients. Front Oncol 2021;11:725292. [Crossref] [PubMed]


