
Circ_0087378 intensifies the malignant behavior of non-small cell lung cancer cells in vitro by facilitating DDR1 via sponging miR-199a-5p
Highlight box
Key findings
• Circ_0087378 is an oncogene for NSCLC.
What is known and what is new?
• Circ_0087378 has been discovered to be involved in the progression of some cancers, and its function varies depending on the types of cancers. However, the expression and function of circ_0087378 in NSCLC has never been identified.
• This paper firstly identified the expression and function of circ_0087378 in NSCLC. It was found that, circ_0087378 silencing attenuated proliferation, colony formation, migration, and invasion of NSCLC cells and promoted its apoptosis. Mechanistically, circ_0087378 might promote the progression of NSCLC by promoting DDR1 via sponging miR-199a-5p.
What is the implication, and what should change now?
• Circ_0087378 exerts oncogenic activity in NSCLC. It may be promising target for the treatment of NSCLC.
Introduction
Non-small cell lung cancer (NSCLC) accounts for over 80% of lung cancer diagnoses (1). Heredity, environment, immunity, and bad habits (especially smoking) are common risk factors for NSCLC (2). The great advances have been achieved in the diagnosis and therapy of NSCLC. However, the prognosis of NSCLC cases is still unfavorable, due to its rapid proliferation and unmanageable metastasis (3). The 5-year survival rate of NSCLC patients is only about 15% (4). For patients with advanced stages, surgical resection is no longer applicable, and non-surgical treatments (such as chemotherapy, immunotherapy and targeted therapy) are commonly used (5). Thus, elucidating the underlying pathogenesis of NSCLC is one of the top priorities in developing effective therapeutic regimens.
Circular RNAs (circRNAs) are a group of highly conserved endogenous molecules with stable covalently closed circular structures. These special structural advantages make circRNAs more useful than linear genes as targets for cancer treatment (6). At present, the function of circRNAs in cancer progression has attracted increasing attention, and researchers are seeking to develop effective drugs for the target treatment of cancer (7). Famously, circRNAs can participate in the post-transcriptional regulation of coding gene expression in cancer by acting as microRNA (miRNA) sponges (8). The significance of circRNAs in the pathogenesis of NSCLC has been established (9,10). CircRNA (circ)_0087378 has been recently identified to be down-regulated in breast cancer; it suppressed the progression of breast cancer by elevating secreted frizzled related protein-1 (SFRP1) expression via sponging miRNA (miR)-1260b (11). However, circ_0087378 has been verified to be up-regulated in esophageal squamous cell carcinoma, associating with poor patient outcomes. Circ_0087378 knockdown can attenuate the malignant development in vitro and in vivo growth of esophageal squamous cell carcinoma cells. Mechanically, circ_0087378 may exert tumor-promoting activities by facilitating the expression of E2F transcription factor 3 (E2F3) via sponging miR-140-3p (12). Thus, the role of circ_0087378 in cancers remains controversial, and more data about the function of circ_0087378 in other tumors are still not available. At present, there are many circRNAs that have been found to regulate the malignancy of NSCLC. For example, circular pleiotrophin (circPTN) and circRNA C190 have been proven to be oncogenic for NSCLC (13,14). Conversely, circ_0002346 and circ_0003176 have been identified to be tumor suppressors for NSCLC (15,16). However, the expression and function of circ_0087378 has never been researched currently. Thus, this paper aimed to elucidate the function of circ_0087378 in NSCLC in order to finding a novel target for NSCLC treatment.
According to our preliminary study, circ_0087378 was discovered to be abundantly expressed in NSCLC cell lines. Therefore, we speculated that circ_0087378 exerts a regulatory role in the pathogenesis and development of NSCLC. We further implemented bioinformatics analysis to explore the molecular mechanisms of circ_0087378 in modulating NSCLC progression. It was found that circ_0087378 contained the binding sites of miR-199a-5p and miR-199a-5p had the binding sites of discoidin domain receptor 1 (DDR1). MiR-199a-5p has been implied to exert tumor suppressor activity in NSCLC (17) and the high expression of DDR1 can intensify the invasiveness of NSCLC cells (18). Therefore, in the present study, we verified whether circ_0087378 regulated the malignant behavior of NSCLC cells in vitro via targeting miR-199a-5p/DDR1. We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-88/rc).
Methods
Cell line and culture
The human normal bronchial epithelial cell line [16HBE (catalogue No. XY-XB-2187)] and human NSCLC cell lines [A549 (catalogue No. XY-XB-1166), NCI-H1299 (catalogue No. XY-XB-1528), NCI-H1975 (catalogue No. XY-XB-1347), HCC827 (catalogue No. XY-XB-1356), and PC9 (catalogue No. XY-XB-1494)] were all purchased from the American Type Culture Collection China agent company (Xuanya Biotechnology, Shanghai, China). The cells were individually fostered in Dulbecco’s modified eagles medium [DMEM; catalogue No. 11995; Solarbio, Beijing, China)] containing 10% fetal bovine serum (FBS; catalogue No. S9030; Solarbio) in sterile culture flasks at 37 ℃ and 5% carbon dioxide (CO2).
Nucleocytoplasmic separation experiment
A549 and NCI-H1299 cells were harvested at about 85% confluence. The subcellular localization of circ_0087378 in A549 and NCI-H1299 cells were researched via a nucleocytoplasmic separation experiment using the nucleocytoplasmic separation kit (catalogue No. SC-003; Invent Biotechnologies, Beijing, China). The operation was implemented according to the kit directions. After the extraction of total RNA in the cytoplasm and nucleus, we detected the level of circ_0087378 in the cytoplasm and nucleus by quantitative real-time polymerase chain reaction (qRT-PCR). U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as the control for the level of circ_0087378 in the nucleus and cytoplasm.
Cell transfection
The circ_0087378 short hairpin RNA (shRNA) (sequence #1 5'-GGCATGAAAGGTGCAAACCCA-3' and sequence #2 5'-GCATGAAAGGTGCAAACCCAA-3') and negative control (NC) (sequence 5'-UUCUCCGAACGUGUCACGUTT-3') plasmids were purchased from GeneChem (Shanghai, China). A549 and NCI-H1299 cells (1×106 cells in 1 mL non-serum DMEM) were fostered into six-well plates. The shRNA and NC plasmids were then transfected into A549 and NCI-H1299 cells based on the directions of Lipofectamine 3000 (catalogue No. L3000-015; Solarbio). These A549 and NCI-H1299 cells were classified as the sh-circ_0087378#1, sh-circ_0087378#2, and sh-NC groups, respectively.
As described above, miR-199a-5p mimic, mimic NC, miR-199a-5p inhibitor, inhibitor NC, protruding clustered DNA (pcDNA)-DDR1 vectors, and empty vectors (GeneChem) were respectively transfected into A549 and NCI-H1299 cells and classified as the miR-199a-5p mimic, miR-NC, anti-miR-199a-5p, anti-NC, pcDNA-DDR1, and vector groups, respectively. Moreover, A549 and NCI-H1299 cells underwent co-transfection with circ_0087378 shRNA (sequence #1) and miR-199a-5p inhibitor (sh-circ_0087378#1 + anti-miR-199a-5p group), or miR-199a-5p mimic and pcDNA-DDR1 vectors (miR-199a-5p mimic + pcDNA-DDR1 group).
Further, pcDNA-circ_0087378 vectors and pcDNA vectors (GeneChem) were individually transfected into A549 cells (named circ_0087378 group and con group).
Following transfection, the A549 and NCI-H1299 cells were fostered for 48 h with DMEM containing 10% FBS at 37 ℃ and 5% CO2.
Cell counting kit-8 (CCK-8) assay
TheA549 and NCI-H1299 cell suspension were prepared by dispersing cells into DMEM (10% FBS) (1×105 cells/mL). After being inoculated into 96-well plates (100 µL cell suspension per well), the cells were grown for 24, 48, and 72 h, respectively, at 37 ℃ and 5% CO2. At specific time points, the CCK-8 solution (10 µL catalogue No. CA1210; Solarbio) treatment was performed on cells for 2 h. The optical density (OD) value was detected by a multi-well microplate reader (Multiskan FC; Thermo Fisher Scientific, Waltham, MA, USA) at 450 nm to evaluate the proliferative ability.
Colony formation assay
A549 and NCI-H1299 cell suspension (1×103 cells/mL) was fostered into six-well plates with 1 mL per well. The plates were maintained for 14 days at 37 ℃ and 5% CO2. The medium was replaced at a 3-day interval. On the 14th day, the cells were washed twice with phosphate-buffered saline (PBS; catalogue No. P1010; Solarbio). Next, the cells were sequentially subjected to fixation using 4% (catalogue No. P1110; Solarbio) paraformaldehyde and stained with 0.1% crystal violet (catalogue No. G1063; Solarbio). The plates were then placed under a light microscope (BX60; Olympus, Tokyo, Japan) to count the colony formation number.
Transwell assay
The Transwell inserts were utilized to research the migration capacity and invasion ability of the A549 and NCI-H1299 cells. Briefly, the A549 and NCI-H1299 cells were prepared into cell suspension by dispersing into DMEM without serum (1×106 cells/mL). The cell suspension with a volume of 200 µL was filled into the upper chambers [pre-coated with or without Matrigel (catalogue No. 356234; Solarbio)]. DMEM with 10% FBS (600 µL) was plated into the lower chambers for induction. The Transwell inserts were then placed at 37 ℃ and 5% CO2 for 24 h. The migrating and invading cells were attached to the lower surface of the chambers. After fixation using 4% paraformaldehyde and staining with 0.1% crystal violet, a light microscope (BX60; Olympus) was employed to observe the migrating and invading cells in five random fields of view.
Flow cytometry
The A549 and NCI-H1299 cells were harvested and washed twice using pre-cooled PBS. The Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (catalogue No. C1062S; Beyotime, Shanghai, China) was recruited to stain the cells. The detection process was performed strictly according to the manufacturer’s directions. Then, the apoptosis of cells was investigated using the FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Dual-luciferase reporter gene assay
Using online miRDB bioinformatics software (http://www.mirdb.org/cgi-bin/custom.cgi), miR-199a-5p was discovered to contain the mutual binding sites of circ_0087378 and DDR1. Based on the mechanism of the circRNAs/miRNAs/messenger (mRNAs) regulatory network, we inferred that circ_0087378 might modulate NSCLC progression by regulating DDR1 via targeting miR-199a-5p. A dual-luciferase reporter gene assay was then performed to monitor the binding between miR-199a-5p and circ_0087378 (or DDR1). The circ_0087378 wild type (WT), circ_0087378 mutant type (MUT), DDR1 WT, and DDR1 MUT sequences were synthesized by GeneChem and cloned into the luciferase reporters according to the manufacturer’s instructions. The miR-199a-5p mimic or mimic NC-transfected A549 and NCI-H1299 cells were then co-transfected by the luciferase reporters according to the manufacturer’s directions. Following co-transfection, the A549 and NCI-H1299 cells were fostered for 48 h with DMEM containing 10% FBS. We utilized the Dual-Luciferase Reporter Gene Assay Kit (catalogue No. RG027; Beyotime, Shanghai, China) to monitor the luciferase activity. The renilla luciferase activity was used as the control to normalize the relative luciferase activity.
RNA pull-down assay
A549 and NCI-H1299 cells were harvested and lysed by cell lysate (catalogue No. P0013; Beyotime) to collect the supernatant. The supernatant (50 µL) was incubated for 4 h with biotin-labeled circ_0087378 probes or oligo probes (GeneChem) at room temperature. An equal volume of the supernatant without any treatment was used as the input. For the capture of circ_0087378-miRNAs complexes, streptavidin-coupled Dynabeads (catalogue No. 65001; Qianxi Biotechnology, Shanghai, China) were added to incubate for 12 h. The beads were then collected via centrifugation and treated by RNA immunoprecipitation (RIP) wash buffer (with proteinase K) (catalogue No. QY-Xz228; Qiao Yu Biotechnology, Shanghai, China) to collect the RNA complexes. The complexes were treated with TRIzol (catalogue No. 15596-026; Boster, Wuhan, China) to extract RNA. qRT-PCR was implemented to analyze the level of miR-199a-5p pulled down by the circ_0087378 probes.
qRT-PCR
Total RNA in cells was harvested by TRIzol treatment (catalogue No. 15596-026; Boster) according to the manufacturer’s directions. The total RNA sample (5 µg) was collected for reverse transcription to synthesize the complementary DNA (cDNA) of circRNA and mRNA using the Reverse Transcriptase Kit (catalogue No. YZM247; Yuncui Biotechnology, Wuxi, China). A MicroRNA Reverse Transcription Kit (catalogue No. MT0006; Biolab Technology, Beijing, China) was utilized for the synthesis of miRNA cDNA.
Following reverse transcription, PCR was executed on the ABI 7500 fast real-time PCR System (Applied Biosystems, Foster City, CA, USA) according to the following parameters: 95 ℃ for 10 s, then 58 ℃ for 10 s, and finally 72 ℃ for 10 s. The procedure was cycled 40 times. The primers were as follows: circ_0087378, forward, 5'-ACTGGTTTCGTTAGCCCTCGTA-3', reverse, 5'-CGTTAGTGGTGGTGCCGTATCT-3'. miR-1224-3p, forward, 5'-GTCGTATCCAGTGCAGGGT-3', reverse, 5'-AACAAGCCCCACCTCCTCTC-3'. miR-197-3p, forward, 5'-GTCGTATCCAGTGCAGGGT-3', reverse, 5'-AACAAGTTCACCACCTTCTCCAC-3'. miR-1298-5p, forward, 5'-GTCGTATCCAGTGCAGGGT-3', reverse, 5'-AACACGCTTCATTCGGCTGTC-3'. miR-140-3p, forward, 5'-GTCGTATCCAGTGCAGGGT-3', reverse, 5'-TATCCTTGTTCACGACTCCTTCAC-3'. miR-199a-5p, forward, 5'-GTCGTATCCAGTGCAGGGT-3', reverse, 5'-AACAGACCCAGTGTTCAGACT-3'. DDR1, forward, 5'-GAACACGGACGAGGACATG-3', reverse, 5'-CGGCAACGAGGGAGAAAGG-3'. Solute carrier 25A23 (SLC25A23), forward, 5'-GGCCAGATAGCCAGTTACCC-3', reverse, 5'-GGGACAGGATGTGACGTAGC-3'. N-alpha-acetyltransferase 40 (NAA40), forward, 5'-TGCGGATCCAATGGGGAGAAAGTCAAGC-3', reverse, 5'-TTGCTCGAGCCGTGGCAGCAGCCACCACA-3'. Zinc finger protein 763 (ZNF763), forward, 5'-AGAGCCAGATACGGCACCACCA-3', reverse, 5'-GCGTCCCTGTTTCCACTCCATT-3'. Myelin regulatory factor gene (MYRF), forward, 5'-GGTTTCGTTAGCCCTCGTA-3', reverse, 5'-GTCCCTGTTTCCACTCCATTC-3'. U6, forward, 5'-CTCGCTTCGGCAGCACA-3', reverse, 5'-AACGCTTCACGAATTTGCGT-3'; β-actin, forward, 5'-ATGTGGCCGAGGACTTTGATT-3', reverse, 5'-AGTGGGGTGGCTTTTAGGATG-3'. The relative expression of genes was performed according to the 2−ΔΔCt method and normalized to β-actin (for circ_0087378 and mRNAs) and U6 (for miRNAs), respectively.
Western blot
The total protein in cells was extracted by incubating cells with radioimmunoprecipitation assay (RIPA) lysis buffer (catalogue No. P0013B; Beyotime). After determining the concentration, the total protein samples with 30 µg were subjected to 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis for separation. Next, the separated proteins were transferred onto polyvinylidene difluoride (PVDF; catalogue No. FFP33; Beyotime) membranes, and then blocked using 5% non-serum milk (catalogue No. D8340; Solarbio). Rabbit anti-DDR1 (1:1,000; catalogue No. PAB2869; AmyJet Scientific, Wuhan, China) and anti-β-actin (1:1,000; catalogue No. A-AJ1089a; AmyJet Scientific) were respectively added to probe the proteins overnight at 4 ℃. The proteins were then treated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:2,000; catalogue No. ab6721; Abcam, Cambridge, UK) for 2 h at room temperature. The PVDF membranes were then washed three times washing using Tris-buffered saline with 0.1% Tween 20 (TBST; catalogue No. T1085; Solarbio). The specific protein blots were developed via treatment with enhanced chemiluminescence reagent (catalogue No. PE0010; Solarbio) and qualified by Image J software (version 1.43; NIH, USA).
Statistical analysis
Statistical analysis was performed using the SPSS 19.0 software (IBM, Armonk, NY, USA). Data was processed into the form of mean ± standard deviation. The two-tailed Student’s t-test was applied for the data comparison between two groups. One-way analysis of variance with Tukey’s post hoc test was utilized for the data comparison in multiple groups (at least three groups). P<0.05 predicted a statistically significant difference. All experiments were repeated in triplicates in order to confirm the reproducibility of the results. Data described biological replicates.
Results
circ_0087378 was abundantly expressed in NSCLC cells
qRT-PCR showed that circ_0087378 was abundantly expressed in NSCLC cell lines (A549, NCI-H1299, NCI-H1975, HCC827, and PC9), compared to the normal bronchial epithelial cell line (16HBE) (P<0.001) (Figure 1A). The nucleocytoplasmic separation experiment demonstrated that circ_0087378 was mainly expressed in the cytoplasm of A549 and NCI-H1299 cells (Figure 1B).

Knockdown of circ_0087378 repressed the malignant phenotype of NSCLC cells in vitro
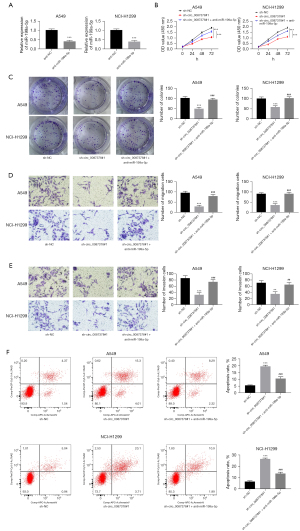
To explore the function of circ_0087378 in NSCLC cell in vitro malignant phenotype, A549 and NCI-H1299 cells were subjected to circ_0087378 shRNA and shNC transfection. circ_0087378 shRNA transfection (sh-circ_0087378#1 and sh-circ_0087378#2 groups) significantly reduced the level of circ_0087378 in A549 and NCI-H1299 cells, relative to the sh-NC group (P<0.001) (Figure 2A).

The malignant phenotype of cells in each group was then monitored. Compared to the sh-NC group, A549 and NCI-H1299 cells of the sh-circ_0087378#1 and sh-circ_0087378#2 groups displayed the attenuated proliferation, colony formation, migration, and invasion capacity, as well as the intensified apoptosis ability (P<0.001) (Figure 2B-2F). Thus, circ_0087378 knockdown repressed the malignant phenotype of NSCLC cells in vitro.
circ_0087378 could directly suppress miR-199a-5p by acting as a sponge
Through bioinformatics online prediction, this study selected the potential target miRNAs of circ_0087378. Finally, we selected five miRNAs that have contained more binding sites of circ_0087378 and have been identified to be down-modulated in lung cancer, including miR-1224-3p, miR-197-3p, miR-1298-5p, miR-140-3p and miR-199a-5p. Then we overexpressed circ_0087378 in A549 cells by pcDNA-circ_0087378 vectors, and detected the expression of the five miRNAs. It was found that circ_0087378 overexpression had the most pronounced down-regulation of miR-199a-5p among the five miRNAs (Figure S1). Thus, this study selected miR-199a-5p as the subject.
Subsequently, this paper transfected A549 and NCI-H1299 cells by circ_0087378 shRNA. As shown in Figure 3A, the loss of circ_0087378 intensified the expression of miR-199a-5p in A549 and NCI-H1299 cells (P<0.001). The dual-luciferase reporter gene assay showed that, miR-199a-5p up-modulation diminished the luciferase activity of the circ_0087378-WT reporters (rather than circ_0087378-MUT reporters) in A549 and NCI-H1299 cells (P<0.001) (Figure 3B). According to the RNA pull-down assay, circ_0087378 probes were able to pull down more miR-199a-5p than the oligo probes (P<0.001) (Figure 3C). These results suggested that circ_0087378 could repress miR-199a-5p by acting as a sponge.

Silencing of miR-199a-5p abrogated the inhibition of circ_0087378 loss on the malignant phenotype of NSCLC cells in vitro
qRT-PCR showed that, miR-199a-5p inhibitor transfection (anti-miR-199a-5p group) effectively reduced miR-199a-5p expression in A549 and NCI-H1299 cells compared to inhibitor NC transfection (anti-NC group) (P<0.001) (Figure 4A). The weakened proliferation, colony formation, migration, and invasion abilities, and the intensified apoptosis capacity, were found in A549 and NCI-H1299 cells of the sh-circ_0087378#1 group, when relative to the sh-NC group (P<0.01 and P<0.001). However, miR-199a-5p inhibitor abrogated these effects of circ_0087378 silencing on the proliferation, colony formation, migration, invasion, and apoptosis abilities (P<0.01 and P<0.001) (Figure 4B-4F). Therefore, miR-199a-5p silencing counteracted the repressive role of circ_0087378 loss on the malignant phenotype of NSCLC cells in vitro.
DDR1 was directly repressed by miR-199a-5p
This paper predicted the potential target mRNAs of miR-199a-5p via online bioinformatics analysis. Among these mRNAs, we selected the five mRNAs that have more binding sites of miR-199a-5p and have been shown to be up-regulated in cancers. As a result, DDR1, SLC25A23, NAA40, ZNF763 and MYRF were selected. This study was then overexpressed miR-199a-5p in A549 cells by miR-199a-5p mimic transfection to detect the effect of miR-199a-5p on the expression of the five target mRNAs. As a result, miR-199a-5p overexpression showed the stronger inhibitory effect on DDR1 mRNA expression than on the others (Figure S2). Therefore, DDR1 was selected to be the subject in this research.
Based on dual-luciferase reporter gene assay, miR-199a-5p up-modulation reduced the luciferase activity of the DDR1-WT reporters (rather than the DDR1-MUT reporters) in A549 and NCI-H1299 cells (P<0.001) (Figure 5A). miR-199a-5p up-modulation suppressed the expression of DDR1 protein in A549 and NCI-H1299 cells (P<0.01) (Figure 5B). Simultaneously, the A549 and NCI-H1299 cells of the sh-circ_0087378#1 group expressed lower DDR1 protein, compared to the sh-NC group and the sh-circ_0087378#1 + anti-miR-199a-5p group (P<0.01 and P<0.001) (Figure 5C). Hence, DDR1 expression was directly repressed by miR-199a-5p.

DDR1 counteracted the inhibition of miR-199a-5p on the malignant phenotype of NSCLC cells
As shown in Figure 6A, higher DDR1 protein was found in A549 and NCI-H1299 cells of the pcDNA-DDR1 group, relative to the vector group (P<0.05 and P<0.01). Thus, the pcDNA-DDR1 vectors were effectively transfected into A549 and NCI-H1299 cells. Functional studies showed that, relative to the miR-199a-5p mimic group, the proliferation, colony formation, migration, and invasion abilities were potentiated in A549 and NCI-H1299 cells of the miR-NC and miR-199a-5p mimic + pcDNA-DDR1 groups; however, the increased apoptosis capacity of A549 and NCI-H1299 cells in the miR-199a-5p mimic group was exhibited, when relative to the miR-NC and miR-199a-5p mimic + pcDNA-DDR1 groups (P<0.05, P<0.01 and P<0.001) (Figure 6B-6F). These results implied that, DDR1 counteracted the inhibitory role of miR-199a-5p on the malignant phenotype of NSCLC cells in vitro.

Discussion
In the present study, circ_0087378 was identified as an oncogene in NSCLC. It intensified the malignant behavior of NSCLC cells in vitro by facilitating DDR1 via acting as a miR-199a-5p sponge. At present, multiple circRNAs are expected to be promising targets for NSCLC treatment. For instance, circ_0041268, circ_0020123, and circ_0017639 have been verified to be tumor promoters. They facilitate NSCLC progression by enhancing the expression of downstream coding genes via acting as sponges of their target miRNAs (19-21). On the contrary, circ-PLCD1, circ_0008797, and circPTK2 are regarded as tumor suppressors. These circRNAs attenuate the malignant behavior of tumor cells via sponging the miRNAs/mRNAs axis (22-24). However, the influence of circ_0087378 on NSCLC remains unclear. In the present work, circ_0087378 was found to exert cancer-promoting activity in NSCLC; it intensified the malignant behavior of NSCLC cells in vitro by sponging the miR-199a-5p/DDR1 axis.
In the current research, miR-199a-5p was confirmed as a target of circ_0087378 and it was a tumor suppressor in NSCLC. miR-199a-5p up-modulation could attenuate the proliferation, colony formation, migration, and invasion of NSCLC cells in vitro. Conversely, miR-199a-5p overexpression could intensify the apoptosis of NSCLC cells in vitro. Interestingly, miR-199a-5p silencing abrogated the inhibition of circ_0087378 knockdown on the malignant phenotype of NSCLC cells in vitro. miR-199a-5p has been identified to exert tumor suppressor activity in hepatocellular carcinoma. It’s up-modulation represses the proliferative capacity and induces the cell-cycle arrest of hepatocellular carcinoma cells (25). Also, aberrantly reduced miR-199a-5p has been discovered in bladder cancer clinical specimens, relating to the adverse survival of patients (26). Similarly, the low expression of miR-199a-5p has been revealed in the tumor specimens of laryngeal cancer cases. The up-modulated miR-199a-5p expression restrains the proliferation, invasion, and migration of laryngeal cancer cells but triggers their apoptosis (27). miR-199a-5p overexpression can intensify the chemotherapy sensitivity, weaken the proliferation and invasion, and trigger the apoptosis of NSCLC cells (28,29). Moreover, elevated miR-199a-5p expression can restrain the proliferation and cell cycle progression of NSCLC cells (17). Similarly, the present research revealed the tumor suppressor role of miR-199a-5p in the malignant behavior of NSCLC cells in vitro. Moreover, circ_0087378 exerted its promoting role in the malignant behavior of NSCLC cells in vitro by sponging miR-199a-5p.
DDR1 is a collagen receptor that exerts tyrosine kinase activity. It can intensify immune exclusion by enhancing the alignment of collagen fibers (30). DDR1 is a key driver in the acquisition of mesenchymal and aggressive phenotype in pancreatic ductal adenocarcinoma cells (31). The over- expression of DDR1 is a predictor of poor clinical outcomes in pancreatic ductal adenocarcinoma patients (32). Moreover, DDR1 expression in gastric cancer is positively correlated with unfavorable outcomes in patients. The silencing of DDR1 represses the in vitro migration, invasion, and angiogenesis of gastric cancer cells, but also restrains the formation of lymphatic vessels in vivo to reduce the risk of lymph node and liver metastases in mice (33). In terms of breast cancer, DDR1 silencing can inhibit the migration and survival of tumor cells (34). Available data has also shown that DDR1 is more frequently expressed in invasive NSCLC. DDR1 is conducive to facilitating the motility and invasiveness of NSCLC cells (18). DDR1 up-modulation can exacerbate epithelial-mesenchymal transition in NSCLC cells (35). Herein, DDR1 was verified as a target of miR-199a-5p. DDR1 up-modulation counteracted the inhibition of miR-199a-5p on the malignancy of NSCLC cells in vitro. Taken together, circ_0087378 might intensify the malignant behavior of NSCLC cells in vitro by facilitating DDR1 via acting as a miR-199a-5p sponge. Thus, circ_0087378 may be a novel target for the clinical treatment of NSCLC in the future. The suppression of circ_0087378 may help improve the prognosis of NSCLC patients. Further, this paper provides new insights into the understanding of the etiology of NSCLC. Of course, safe and effective drugs to inhibit circ_0087378 expression still need to be discovered. These challenges still need to be overcome in the clinical application of circ_0087378.
This study has limitations. Firstly, in vivo study with animals is preferable, and it will be conductive to the NSCLC treatment with circ_0087378 as the target. Secondly, the correlations between circ_0087378 and NSCLC staging, degree of differentiation, lymphatic metastasis and survival prognosis should be better researched by collecting clinical data. Further, to show that interfering with the circ_0087378/mir199/DDR1 pathway leads to epithelial mesenchymal transition (EMT) alteration to alter NSCLC cell malignant phenotype, it would be desirable to perform EMT-related experiments. However, these studies cannot currently be carried out due to laboratory limitations and this will be the focus of our future research.
Conclusions
This study in vitro experiments and identified circ_0087378 as an oncogene in NSCLC for the first time. circ_0087378 knockdown suppressed the malignancy of NSCLC cells in vitro. Mechanically, circ_0087378 might potentiate the malignant development of NSCLC cells in vitro by facilitating DDR1 via acting as miR-199a-5p sponge. Thus, circ_0087378 may be a novel target for the clinical treatment of NSCLC in the future.
Acknowledgments
Funding: The study was supported by The 7th Wuhan Young and Middle-aged Backbone Talent of Medical Training Project 2019 (2019 No. 87, to Guoliang Pi).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-88/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-88/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-88/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-88/coif). GP reports that the study was supported by The 7th Wuhan Young and Middle-aged Backbone Talent of Medical Training Project 2019 (2019 No. 87, to Guoliang Pi). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lim JU, Yeo CD. Update on adjuvant therapy in completely resected NSCLC patients. Thorac Cancer 2022;13:277-83. [Crossref] [PubMed]
- Liang H, Peng J. LncRNA HOTAIR promotes proliferation, invasion and migration in NSCLC cells via the CCL22 signaling pathway. PLoS One 2022;17:e0263997. [Crossref] [PubMed]
- Guo K, Ma Z, Zhang Y, et al. HDAC7 promotes NSCLC proliferation and metastasis via stabilization by deubiquitinase USP10 and activation of β-catenin-FGF18 pathway. J Exp Clin Cancer Res 2022;41:91. [Crossref] [PubMed]
- Li Y, Shen Y, Xie M, et al. LncRNAs LCETRL3 and LCETRL4 at chromosome 4q12 diminish EGFR-TKIs efficiency in NSCLC through stabilizing TDP43 and EIF2S1. Signal Transduct Target Ther 2022;7:30. [Crossref] [PubMed]
- Li F, Ye P, Cai P, et al. Prevalence of targeted therapy-related genetic variations in NSCLC and their relationship with clinicopathological characteristics. PLoS One 2022;17:e0262822. [Crossref] [PubMed]
- Ma J, Li Q, Li Y. CircRNA PRH1-PRR4 stimulates RAB3D to regulate the malignant progression of NSCLC by sponging miR-877-5p. Thorac Cancer 2022;13:690-701. [Crossref] [PubMed]
- Zhu B, Ke L, Li P, et al. CircACC1 Promotes NSCLC Proliferation via miR-29c-3p/MCL-1 Signaling Pathway. Front Genet 2022;12:798587. [Crossref] [PubMed]
- Ge L, Tan W, Li G, et al. Circ_0026134 promotes NSCLC progression by the miR-3619-5p/CHAF1B axis. Thorac Cancer 2022;13:582-92. [Crossref] [PubMed]
- Jin J, Xu B, Hu Z, et al. Circular RNA circTADA2A promotes the proliferation, invasion, and migration of non-small cell lung cancer cells via the miR-450b-3p/HMGN5 signaling pathway. Transl Cancer Res 2022;11:242-51. [Crossref] [PubMed]
- Liu X, Chen Z, Wu Y, et al. Circ_0078767 Inhibits the Progression of Non-Small-Cell Lung Cancer by Regulating the GPX3 Expression by Adsorbing miR-665. Int J Genomics 2022;2022:6361256. [Crossref] [PubMed]
- Yuan C, Zhou L, Zhang L, et al. Identification and integrated analysis of key differentially expressed circular RNAs in ER-positive subtype breast cancer. Epigenomics 2019;11:297-321. [Crossref] [PubMed]
- Wang J, Wang Q, Gong Y, et al. Knockdown of circRNA circ_0087378 Represses the Tumorigenesis and Progression of Esophageal Squamous Cell Carcinoma Through Modulating the miR-140-3p/E2F3 Axis. Front Oncol 2021;10:607231. [Crossref] [PubMed]
- Guo S, Mao Z, Zhang G, et al. CircPTN promotes angiogenesis via the MiR-595/LYRM5 signaling pathway in non-small cell lung cancer. Transl Cancer Res 2022;11:519-29. [Crossref] [PubMed]
- Ishola AA, Chien CS, Yang YP, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res 2022;82:75-89. [Crossref] [PubMed]
- Wang W, Lin Y, Zhang G, et al. circ_0002346 Suppresses Non-Small-Cell Lung Cancer Progression Depending on the Regulation of the miR-582-3p/STXBP6 Axis. Int J Genomics 2021;2021:1565660. [Crossref] [PubMed]
- Yang F, Pei Y, Xu W, et al. hsa_circ_0003176 Suppresses the Progression of Non-Small-Cell Lung Cancer via Regulating miR-182-5p/RBM5 Axis. Dis Markers 2022;2022:8402116. [Crossref] [PubMed]
- Li Y, Wang D, Li X, et al. MiR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J Cancer 2019;10:2472-9. [Crossref] [PubMed]
- Yang SH, Baek HA, Lee HJ, et al. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung carcinomas. Oncol Rep 2010;24:311-9. [PubMed]
- Yang W, Wu L, Jin M. Hsa_circ_0041268 promotes NSCLC progress by sponging miR-214-5p/ROCK1. J Clin Lab Anal 2022;36:e24262. [Crossref] [PubMed]
- Fan Y, Wang Q, Shi M, et al. Circ_0020123 promotes NSCLC tumorigenesis via up-regulating KIAA1522 expression through miR-940. Cell Cycle 2022;21:894-907. [Crossref] [PubMed]
- Zhang HB, Qiu XM, Zhang YC, et al. Circ_0017639 facilitates proliferative, migratory, and invasive potential of non-small cell lung cancer (NSCLC) cells via PI3K/AKT signaling pathway. Bioengineered 2022;13:1590-601. [Crossref] [PubMed]
- Si J, Jin J, Sai J, et al. Circular RNA circ-PLCD1 functions as a tumor suppressor in non-small cell lung cancer by inactivation of PI3K/AKT signaling pathway. Hum Cell 2022;35:924-35. [Crossref] [PubMed]
- Abuduwaili K, Zhu X, Shen Y, et al. circ_0008797 attenuates non-small cell lung cancer proliferation, metastasis, and aerobic glycolysis by sponging miR-301a-3p/SOCS2. Environ Toxicol 2022;37:1697-710. [Crossref] [PubMed]
- Wang Y, Wu Y, Xie S. CircPTK2 inhibits cell cisplatin (CDDP) resistance by targeting miR-942/TRIM16 axis in non-small cell lung cancer (NSCLC). Bioengineered 2022;13:3651-64. [Crossref] [PubMed]
- Liu P, Xia P, Fu Q, et al. miR-199a-5p inhibits the proliferation of hepatocellular carcinoma cells by regulating CDC25A to induce cell cycle arrest. Biochem Biophys Res Commun 2021;571:96-103. [Crossref] [PubMed]
- Ma X, Wen Y, Wang Y, et al. Linc00662 plays an oncogenic role in bladder cancer by sponging miR-199a-5p. Am J Transl Res 2021;13:12673-83. [PubMed]
- Li DJ, Wang X, Yin WH, et al. MiR-199a-5p suppresses proliferation and invasion of human laryngeal cancer cells. Eur Rev Med Pharmacol Sci 2020;24:12200-7. [PubMed]
- Jin Y, Wang H, Zhu Y, et al. miR-199a-5p is involved in doxorubicin resistance of non-small cell lung cancer (NSCLC) cells. Eur J Pharmacol 2020;878:173105. [Crossref] [PubMed]
- Yang X, Zheng Y, Tan J, et al. MiR-199a-5p-HIF-1α-STAT3 Positive Feedback Loop Contributes to the Progression of Non-Small Cell Lung Cancer. Front Cell Dev Biol 2021;8:620615. [Crossref] [PubMed]
- Sun X, Wu B, Chiang HC, et al. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature 2021;599:673-8. [Crossref] [PubMed]
- Deng J, Kang Y, Cheng CC, et al. DDR1-induced neutrophil extracellular traps drive pancreatic cancer metastasis. JCI Insight 2021;6:e146133. [Crossref] [PubMed]
- Huo Y, Yang M, Liu W, et al. High expression of DDR1 is associated with the poor prognosis in Chinese patients with pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res 2015;34:88. [Crossref] [PubMed]
- Yuge R, Kitadai Y, Takigawa H, et al. Silencing of Discoidin Domain Receptor-1 (DDR1) Concurrently Inhibits Multiple Steps of Metastasis Cascade in Gastric Cancer. Transl Oncol 2018;11:575-84. [Crossref] [PubMed]
- Wang S, Xie Y, Bao A, et al. Nilotinib, a Discoidin domain receptor 1 (DDR1) inhibitor, induces apoptosis and inhibits migration in breast cancer. Neoplasma 2021;68:975-82. [Crossref] [PubMed]
- Miao L, Zhu S, Wang Y, et al. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung cancer and promotes cell invasion via epithelial-to-mesenchymal transition. Med Oncol 2013;30:626. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


