
Inhibition of BPHL inhibits proliferation in lung carcinoma cell lines
Highlight box
Key findings
• We found that the knockdown of biphenyl hydrolase-like (BPHL) gene expression by short hairpin RNA (shRNA) leads to a decrease in proliferation, colony formation, and metastasis and an increase in apoptosis in lung adenocarcinoma cell lines in vitro.
What is known and what is new?
• The BPHL gene is located on chromosome 6p25, a locus clustered with all identified serine hydrolases so far. The role of BPHL in lung cancer biology has not been elucidated.
• We found the inhibition of BPHL expression in lung adenocarcinoma cell lines led to decreased cell proliferation, increased apoptosis, decreased colony formation, decreased metastasis, and altered cell cycle phase distribution. BPHL knockdown tumor cells grew more slowly and weighed less in vivo.
What is the implication, and what should change now?
• Our study found that BPHL promoted lung cancer carcinogenesis, which warrants further clinical and mechanistic studies.
Introduction
According to our previous study, the metastasis-associated gene 1 (MAT1) protein, an important regulator for tumor metastasis, promotes tumor metastasis in non-small cell lung cancer (NSCLC). The results from our earlier study demonstrated that small interfering RNA (siRNA) 125b inhibited tumor metastasis in vitro and reversed the enhancement effect of MTA1 on cell migration (1).
Using a bioinformatics tool kit, we analyzed the lung carcinoma data in The Cancer Genome Atlas (TCGA) database. We found that miR-125b affected the migration of NSCLC via biphenyl hydrolase-like (BPHL), which is a gene located on chromosome 6p25, a locus clustered with all identified serine hydrolases so far (2). However, except for renal carcinoma (3,4), leukemia (5), lower-grade gliomas (6), and multiple sclerosis (7) patients the role of BPHL in cancer biology has not been elucidated. At present, there are no studies reporting on its role in lung cancer.
In this study, we identified BPHL as a lung cancer-associated gene through a bioinformatics analysis of TCGA database. We observed that the loss of function in BPHL led to significant decreases in proliferation and migration in the human lung adenocarcinoma (LUAD) cell lines, A549 and NCI-H1299. The tumor implantation study recapitulated the effect of BPHL knockdown that was observed in the in vitro studies. We present this article in accordance with the ARRIVE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-225/rc).
Methods
Gene expression and disease correlation analysis to identify BPHL
We downloaded the RNA-sequencing (RNAseq) data obtained from both the tumor and adjacent normal tissues of 57 cases as well as the pathology data in the LUAD category in TCGA database. The data were normalized using the Trimmed Mean of M-values (TMM). The gene expression values were presented as Log2 in scale, and the threshold was set as ±1 to estimate the discrete distribution. The BPHL gene was identified as a top hit (Figure 1 and Table 1) (8-13). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).

Table 1
| Gene ID | Gene symbol | Transcription quantity | Articles on PubMed | Novoseek disease relationships for the gene | MalaCards disease relationships for the gene |
|---|---|---|---|---|---|
| 670 | BPHL | 2 | 28 | 3 | 0 |
Genes related to the cancers of this study in which the function and clinical significance have been reported in the literature, multiple-pass transmembrane protein genes, and genes that were annotated unclearly (such as genes annotated with an open reading frame) were excluded. Also, combined with the gene disease database, a final gene list was obtained and then randomly reduced to determine the final gene list for analysis. TCGA, The Cancer Genome Atlas; BPHL, biphenyl hydrolase-like.
Lentivector expressing ribonucleic acid interference (RNAi) for BPHL
Multiple 19–21 nt RNA sequences targeting BPHL were designed based on the principles of designing RNAi. After assessment using design software, ATCCGAGATGTTTCCAAAT with a GC% content of 36.8% was chosen as the targeting site, which was shuttled into the GV115 lentivector, followed by virus packaging.
Screening human lung carcinoma cell lines that express BPHL
Three common human lung carcinoma cell lines, A549, NCI-H1975, and NCI-H1299 were chosen to test for their BPHL gene expression using quantitative polymerase chain reaction (qPCR). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene, and the 5' and 3' primers were CCATTTCAGCACCTCGGTA and CAGCATCTTTTGCATCCCT, respectively, with an amplicon size of 266 bp (Figure 2A). The screened lung cancer cell line was used as a follow-up experimental cell line.

NCI-H1299 and A549 cells were cultured in RPMI 1640 + 10% FBS and transduced with the GV115-RNAi-BPHL and control lentiviruses. At 72 hours post-transduction, the cells displayed good viability. Detect the transfection effect of cells as measured by the percentage of fluorescence-positive cells. Finally, the expression of mRNA and protein was detected by qPCR and Western blot.
Assessing the effect BPHL knockdown on the proliferation, apoptosis, colony formation, metastasis, and cell cycle of cancer cells
The cells were counted with an MTT (Genview, Cat. No. JT343) assay and Celigo (Nexcelom) using a 96-well plate (100 µL/well). A total of 200 cells were seeded in each well and incubated for 120 hours at 37 °C in a humidified atmosphere with 5% CO2 (14,15). Images were taken every 24 hours to evaluate the effect of BPHL knockdown on cell proliferation.
Cell apoptosis, as measured by caspase 3/7 (Promege, Cat. No. JG8091) activity, was tested according to the manufacturer’s recommended protocol. Briefly, 10,000 cells in 100 µL were seeded into a 96-well plate and incubated for 36 hours at 37 °C in a humidified atmosphere with 5% CO2. The caspase 3/7 activity was then assayed (16).
Colony formation was evaluated in a six-well tissue culture plate. A total of 800 cells were seeded in 2 mL of culture media and were cultured for 5 days at 37 °C in a humidified atmosphere with 5% CO2. On day 5, 1,000 µL of crystal violet (Sangon Biotech, Cat. No. CB0331) was added to stain cells for 20 min, followed by imaging and colony counting. The number of colonies containing ≥50 cells was counted using a microscope (17,18). The experiment was performed with three replicates for each cell line.
Cell migration toward the serum was performed in a Transwell chamber (Corning, Cat. No. 3422) to investigate the role of BPHL on cell migration. A 24-well plate with an upper chamber volume of 100 µL and a lower chamber volume of 600 µL was used (corning). A total of 100,000 cells were seeded into the upper chamber and incubated for 20 hours, followed by counting cells in the lower chamber. For quantification, the cells in the lower compartment were stained with Giemsa stain (Sigma, Cat. No. 32884) and counted in five randomly chosen fields (×200) under a microscope (19). The experiment was performed with three replicates.
The cell cycle was examined by PI staining (Sigma, Cat. No. P4170) of the DNA in cells followed by measurement of the PI signal by fluorescence activated cell sorter (FACS) (Millipore, Billerica, MA, USA). The cells were seeded into a six-well plate (2 mL/well) and cultured for 4 days to reach 80% confluence (20).
Effect of BPHL knockdown on tumor growth in a mouse model of tumor Implantation in nude mice
A549 cells were cultured in a 10 mL tissue culture flask in RPMI 1640 + 10% FBS. At 72 hours post-lentiviral transduction of GV115-BPHL-RNAi or control virus, microscope inspection was performed to confirm the transduction efficiency. The cells were then cultured for an additional 120 hours before being implanted into nude mice.
Twenty 4-week-old nude Balb/c mice were purchased from Shanghai Lingchang Biotechnology Co., Ltd. (China). A total of 4E6 BPHL RNAi-expressing or control A549 cells in 200 µL of phosphate buffered saline (PBS) were injected subcutaneously into the right dorsal skin of the mice. All mice were maintained at 21–22 °C with a relative humidity of 60% and a light/dark cycle every 12 hours. The health condition and tumor growth were monitored over 4 weeks following inoculation of the tumor cells. Tumor diameters were determined in two perpendicular dimensions using a caliper twice a week. Tumor volume was calculated according to the following equation: volume =½ (length × width2). The tumor became detectable starting in week 3 post-inoculation. In week 4, all of the mice were taken down and the tumors were excised. A protocol was prepared before the study without registration.
Statistical analysis
SPSS 22 Windows version was used for statistical analysis. All of the experiments were repeated at least three times. A two-sample t-test was used for statistical inference. P<0.05 was considered statistically significant.
Ethical statement
Animal experiments were performed under a project license (No. NFYY-2019-0046) granted by the animal ethic committee of Nanfang hospital, Southern Medical University. All animal experiments described in this study were carried out in accordance with institutional guidelines for the care and use of animals.
Results
BPHL gene expression and disease correlation analysis to identify BPHL
We analyzed TCGA database using bioinformatics methods and found that the BPHL gene is associated with lung cancer (Figure 1 and Tables 1,2), the expression difference in BPHL genes between the cancer and adjacent tissues in TCGA database [data obtained from both the tumor and adjacent normal tissues of 57cases as well as the pathology data in the lung adenocarcinoma (LUAD) category] was demonstrated by the FC (2.253) and P value (8.87E-27). Based on bioinformatics data, we performed a preliminary experiment and found that expression difference in BPHL genes between the cancer and adjacent tissues in the lung adenocarcinoma.
Table 2
| ID | Gene symbol | FC | P value | Total Sample | Sample unchanged | Sample up | Sample down |
|---|---|---|---|---|---|---|---|
| 670 | BPHL | 2.253 | 8.87E-27 | 57 | 24 | 33 | 0 |
The expression difference in BPHL genes between the cancer and adjacent tissues in TCGA database was demonstrated by the FC (the ratio of expression between cancer and adjacent tissues) and P value (the value in the statistical analysis model that determines whether the null hypothesis is met or not). BPHL, biphenyl hydrolase-like; TCGA, The Cancer Genome Atlas; FC, the ratio of expression between cancer and adjacent tissues.
Lentivector-expressing RNAi for BPHL and screening human lung carcinoma cell lines that express BPHL
Three common human lung carcinoma cell lines, A549, NCI-H1975, and NCI-H1299, were chosen to test for their gene expression of BPHL using qPCR. GAPDH was used as a reference gene, NCI-H1299 and A549 cells expressing the highest and lowest BPHL mRNA, respectively, were selected for the subsequent experiments.
NCI-H1299 and A549 were transduced with GV115-RNAi-BPHL lentivirus and a control lentivirus. At 72 hours post-transduction, the cells displayed good viability. The transduction efficiency had reached 80%, as measured by the percentage of fluorescence-positive cells (Figure 2B). mRNA levels were measured by qPCR to confirm the efficiency of the knockdown (Figure 2C). Western blot demonstrated that there was a significant decrease in BPHL at the protein level, as compared to the controls (Figure 2D).
BPHL knockdown leads to decreased tumor growth, colony formation, metastasis, increased apoptosis, and altered cell cycle destruction
After BPHL knockdown with small hairpin RNA (shRNA), we found that NCI-H1299 and A549 cell lines displayed decreased proliferation compared to the controls (Figure 3A,3B). Cell counting results from the MTT assay were consistent with the Celigo cell counting results (Figure 3C). Also, caspase 3/7 activity increased significantly in the NCI-H1299 and A549 cells after shBPHL knockdown (Figure 3D). Additionally, we observed decreased colony formation in the shBPHL knockdown NCI-H1299 and A549 cells (Figure 3E). Our data suggest that BPHL knockdown leads to decreased tumor growth and colony formation and increased apoptosis.
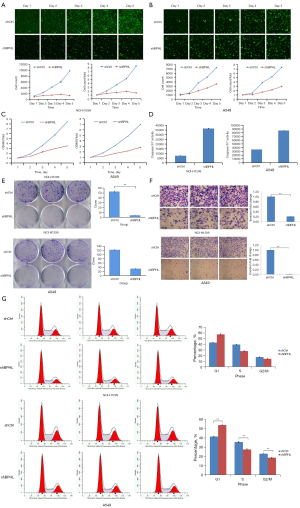
The transmigration assay using a Transwell demonstrated that there were significantly fewer migrating cells in the lower chamber in the BPHL knockdown NCI-H1299 and A549 cells, suggesting that a loss of function in BPHL could lead to decreased tumor metastasis (Figure 3F).
On day 4 post-lentiviral transduction, there were fewer BPHL knockdown NCI-H1299 and A549 cells in the S and G2/M phases but more in the G1 phase compared to the control cells, suggesting that the BPHL gene is correlated with cell cycle distribution. Most BPHL knockdown NCI-H1299 and A549 cells were in the prophase of the cell cycle (Figure 3G). These data indicate that BPHL gene knockdown would negatively affect tumor cell mitosis, leading to impaired cell mitosis.
BPHL knockdown leads to decreased tumor growth in vivo
We also studied the role of BPHL in tumor growth in vivo. We subcutaneously injected tumor cells into nude mice and then monitored their health conditions closely and periodically measured the tumor size (Figure 4A,4B). Tumor cells with BPHL knockdown grew more slowly compared to the control cells, as measured by the tumor size (Figure 4C) and weight (Figure 4D).

Discussion
The BPHL gene is a serine hydrolase with molecular weight of 30 kDa, which shares a certain degree of homology with the enzymes that hydrolyze olychlorinated biphenyl in prokaryotes. Its mRNA has eight exons and seven introns, encoding human biphenyl hydrolase-like (BPHL) enzymes that catalyze the hydrolytic activation of amino acid ester prodrugs of nucleoside analogs, such as valacyclovir and valganciclovir (2,21). The complementary DNA (cDNA) of BPHL was first discovered in the human breast cancer gene library. Kim et al. (22) confirmed the importance of BPHL in designing amino acid ester prodrugs. BPHL has the properties of serine hydrolase, belongs to serine lysine hydrolase, and is an important alpha amino acid hydrolase (23,24). It breaks up the large molecule proteins into short peptides by attacking its peptide bond, and plays an important role in mammalian species, especially in digestion, coagulation, and the complement system. However, our literature search did not yield any previous research regarding the role of BPHL in cancer biology.
In this study, we found that the inhibition of BPHL expression in lung cancer cell lines, NCI-H1299 and A549, led to decreased cell proliferation, increased apoptosis, decreased colony formation, decreased metastasis, and altered cell cycle phase distribution. This illustrates that inhibition of BPHL expression can reduce the malignancy of tumor cells, including reduced proliferation, increased apoptosis, reduced colony formation, and reduced metastasis, and by further experiments, it was found that most of the tumor cells that inhibited BPHL expression stopped in the prophase of mitosis, further confirming that inhibiting BPHL expression would reduce the malignant degree of tumor cells, it causes a decline in pathogenicity and may accelerate death. Furthermore, the Table 1 data showed that the difference of BPHL gene expression between the tumor and the adjacent tissue was significant in the current and future lung adenocarcinoma tumor database, namely that BPHL knockdown tumor cells grew more slowly and weighed less in vivo. These data suggest BPHL potentially promotes proliferation, inhibits apoptosis, and increases colony formation and metastasis in lung cancer.
According to current statistics, there were 19.29 million new cancer cases and 9.96 million cancer deaths in 2020, including 2.26 million new cases of breast cancer and 2.2 million new cases of lung cancer, however, lung cancer is still the world’s leading cause of death, accounting for 1.8 million deaths, or 22.7% of the total, far exceeding other cancer types. China has 4.57 million new cases of cancer, accounting for 23.7% of the global total, and 3 million cancer deaths in China, accounting for 30% of the global total. Lung cancer is one of the malignant tumors with the highest morbidity and mortality in China, with 820,000 new cases of lung cancer in 2020, accounting for 18.0% of the total, leading by far the number of lung cancer deaths, reaching 710,000, it accounts for 23.8% of all cancer deaths in China. Current data show that the overall incidence of lung cancer is on the rise worldwide (25). With the rapid development of medicine, the treatment of lung cancer has also undergone tremendous changes, from the traditional thoracotomy, later, micro-invasive surgery (26-29), systemic chemotherapy, molecular targeted therapy (30), immunotherapy, radiotherapy and so on were gradually developed, but the long-term effect of patients with advanced lung cancer was still not ideal. At present, it is thought that the occurrence of lung cancer may be related to long-term smoking, environment and genetic factors, but the specific mechanism of the occurrence and development of lung cancer is not clear. Therefore, we should go deep into the basic research of the occurrence and development of lung cancer, especially lung adenocarcinoma, fully understand the molecular mechanism of proliferation and metastasis, and search for effective and specific key regulatory genes for the occurrence and development of lung cancer, it is of great value not only in predicting the prognosis of lung adenocarcinoma, but also in improving the therapeutic effect and reducing the mortality of lung adenocarcinoma.
At present, there are no studies on its role in tumor tissues and the mechanism underlying its function is unknown. Our study found that BPHL promoted lung cancer carcinogenesis, which warrants further clinical and mechanistic studies. But this study only completed transfection of shRNA to suppress the expression of the BPHL gene; In order to better understand the mechanism of BPHL-induced carcinogenesis, we will further study the overexpression of BPHL gene in lung adenocarcinoma cell lines, and clarify the function of BPHL oncogene and the cellular pathway of BPHL-induced carcinogenesis, further molecular studies are needed to identify the genetic basis of BPHL target proteins and aberrant expression of BPHL in NSCLC for their possible subsequent development of targeted genes for this gene.
Conclusions
In summary, our findings suggest that the inhibition of BPHL expression in lung cancer cell lines, NCI-H1299 and A549, led to decreased cell proliferation, increased apoptosis, decreased colony formation, decreased metastasis, and altered cell cycle phase distribution. This illustrates that inhibition of BPHL expression can reduce the malignancy of tumor cells,it causes a decline in pathogenicity and may accelerate death. Together, these findings suggest that BPHL can be utilized as a potential diagnostic biomarker for NSCLC progression.
Acknowledgments
Funding: This work was supported by the Project of the Guangdong Provincial Natural Science Foundation of China (No. 2018A0303130200).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-225/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-225/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-225/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-225/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Animal experiments were performed under a project license (No. NFYY-2019-0046) granted by the animal ethic committee of Nanfang hospital, Southern Medical University. All animal experiments described in this study were carried out in accordance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li Y, Chao Y, Fang Y, et al. MTA1 promotes the invasion and migration of non-small cell lung cancer cells by downregulating miR-125b. J Exp Clin Cancer Res 2013;32:33. [Crossref] [PubMed]
- Puente XS, Pendás AM, López-otín C. Structural characterization and chromosomal localization of the gene encoding human biphenyl hydrolase-related protein (BPHL). Genomics 1998;51:459-62. [Crossref] [PubMed]
- Qu Y, Feng J, Wu X, et al. A proteogenomic analysis of clear cell renal cell carcinoma in a Chinese population. Nat Commun 2022;13:2052. [Crossref] [PubMed]
- Grigo K, Wirsing A, Lucas B, et al. HNF4 alpha orchestrates a set of 14 genes to down-regulate cell proliferation in kidney cells. Biol Chem 2008;389:179-87. [Crossref] [PubMed]
- Chu C, Liu BZ, Zhong L, et al. Identification of interaction between BPHL and PML-C. Sichuan Da Xue Xue Bao Yi Xue Ban 2013;44:6-9. [PubMed]
- Yan D, Zhao Q, Du Z, et al. Development and validation of an immune-related gene signature for predicting the radiosensitivity of lower-grade gliomas. Sci Rep 2022;12:6698. [Crossref] [PubMed]
- Kular L, Ewing E, Needhamsen M, et al. DNA methylation changes in glial cells of the normal-appearing white matter in Multiple Sclerosis patients. Epigenetics 2022;17:1311-30. [Crossref] [PubMed]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [Crossref] [PubMed]
- Lund SP, Nettleton D, McCarthy DJ, et al. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol 2012;11:/j/sagmb.2012.11.issue-5/1544-6115.1826/1544-6115.1826.xml.
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288-97. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 2007;23:2881-7. [Crossref] [PubMed]
- Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2008;9:321-32. [Crossref] [PubMed]
- Nabzdyk CS, Chun M, Pradhan Nabzdyk L, et al. Differential susceptibility of human primary aortic and coronary artery vascular cells to RNA interference. Biochem Biophys Res Commun 2012;425:261-5. [Crossref] [PubMed]
- Angius F, Floris A. Liposomes and MTT cell viability assay: an incompatible affair. Toxicol In Vitro 2015;29:314-9. [Crossref] [PubMed]
- Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends Biochem Sci 1997;22:299-306. [Crossref] [PubMed]
- Sun R, Wu J, Chen Y, et al. Down regulation of Thrombospondin2 predicts poor prognosis in patients with gastric cancer. Mol Cancer 2014;13:225. [Crossref] [PubMed]
- Ruan J, Zheng H, Fu W, et al. Increased expression of cathepsin L: a novel independent prognostic marker of worse outcome in hepatocellular carcinoma patients. PLoS One 2014;9:e112136. [Crossref] [PubMed]
- Stagg HW, Bowen KA, Sawant DA, et al. Tumor necrosis factor-related apoptosis-inducing ligand promotes microvascular endothelial cell hyperpermeability through phosphatidylinositol 3-kinase pathway. Am J Surg 2013;205:419-25. [Crossref] [PubMed]
- Bank HL. Rapid assessment of islet viability with acridine orange and propidium iodide. In Vitro Cell Dev Biol 1988;24:266-73. [Crossref] [PubMed]
- Kim I, Song X, Vig BS, et al. A novel nucleoside prodrug-activating enzyme: substrate specificity of biphenyl hydrolase-like protein. Mol Pharm 2004;1:117-27. [Crossref] [PubMed]
- Kim I, Chu XY, Kim S, et al. Identification of a human valacyclovirase: biphenyl hydrolase-like protein as valacyclovir hydrolase. J Biol Chem 2003;278:25348-56. [Crossref] [PubMed]
- Puente XS, López-Otín C. Cloning and expression analysis of a novel human serine hydrolase with sequence similarity to prokaryotic enzymes involved in the degradation of aromatic compounds. J Biol Chem 1995;270:12926-32. [Crossref] [PubMed]
- Lai L, Xu Z, Zhou J, et al. Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase. J Biol Chem 2008;283:9318-27. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Cai K, Ren P, Feng S, et al. Unidirectionally progressive left pneumonectomy & mediastinal lymph node dissection. J THORAC DIS 2013;5:886-91. [PubMed]
- Cai K, Zhao H, Wu H, et al. Unidirectionally progressive resection of left upper pulmonary lobe under video-assisted thoracoscopy. J Thorac Dis 2014;6:1843-7. [PubMed]
- Cai K, Wu H, Ren P, et al. Unidirectionally progressive resection of lower right lung cancer under video-assisted thoracoscopy. J Thorac Dis 2013;5:S310-4. [PubMed]
- Cai K, Feng S, Wu H, et al. Unidirectionally thoracoscopic resection of lingual segment of the left upper pulmonary lobe. J Thorac Dis 2014;6:1358-63. [PubMed]
- Cai KC, Liu DG, Wang YY, et al. Gefitinib maintenance therapy in Chinese advanced-stage lung adenocarcinoma patients with EGFR mutations treated with prior chemotherapy. Neoplasma 2015;62:302-7. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


