
Impact of ramucirumab plus erlotinib on circulating cell-free DNA from patients with untreated metastatic non-small cell lung cancer with EGFR-activating mutations (RELAY phase 3 randomized study)
Highlight box
Key findings
• In this study of patients with EGFR-mutated metastatic non-small cell lung cancer, EGFR-activating mutation allele count was suppressed, total circulating cell-free DNA (cfDNA) concentration increased, and short fragment-sized cfDNA increased with combined ramucirumab and erlotinib treatment.
What is known and what is new?
• The RELAY study demonstrated that ramucirumab plus erlotinib improved progression-free survival compared with placebo plus erlotinib in patients with treatment-naïve EGFR-mutated metastatic non-small cell lung cancer.
• This exploratory liquid biopsy addendum of RELAY investigated whether ramucirumab plus erlotinib affects changes in cfDNA levels and fragment size.
What is the implication, and what should change now?
• These results suggest that the additional anti-tumor effect of ramucirumab on tumor and non-tumor cells may contribute to the progression-free survival benefit observed in the RELAY study.
Introduction
Activating mutations in the epidermal growth factor receptor (EGFR) gene are common drivers of non-small cell lung cancer (NSCLC) (1). The presence of these activating mutations has led to the development of targeted therapy for patients with EGFR mutation-positive NSCLC with small-molecule EGFR tyrosine kinase inhibitors (TKIs) (2,3). Despite TKIs being very effective therapy, eventually many patients will develop treatment resistance and disease progression (4,5). Therefore, new treatment that can prolong and enhance first-line EGFR TKI efficacy is desired.
Cell-free DNA (cfDNA) corresponds to DNA circulating within blood and originates from cell lysis, apoptosis, or necrosis. cfDNA consists predominantly of nucleic acids of hematopoietic origin, but in patients with cancer, cfDNA will also include circulating tumor DNA (ctDNA) derived from tumor cells (6). Tumor-derived material circulating in the blood (i.e., liquid biopsy samples) provides a less invasive alternative to tumor biopsies (7). Digital polymerase chain reaction (dPCR) and next-generation sequencing (NGS) have been used to detect and identify tumor mutational status and gene alterations from ctDNA in plasma (8,9). High concordance for the detection of T790M between tumor biopsies and cfDNA from patients with EGFR mutation-positive NSCLC have been observed using dPCR [83.3% (15/18)] (8) and NGS [76.2% (32/42)] (9), indicating the potential use of liquid biopsy samples for monitoring resistance mechanisms to EGFR TKI treatment. Furthermore, tracking cfDNA actively released from the tumor can be used to detect molecular residual disease, which has been shown to be associated with distant recurrence of disease; molecular residual disease may also provide lead time to disease recurrence (10).
Patterns of cfDNA size vary depending on specific conditions, such as in cancer, in the fetus, or in pregnancy (11-13). Moreover, cfDNA size distribution patterns could be useful diagnostic and prognostic markers for cancer. Research has recently demonstrated that cfDNA fragments originating from tumor cells are shorter than those from non-tumor cells (14). Generally, ctDNA fragments aggregate with cfDNA fragment lengths <150 base pairs (bp), with the median cfDNA strand length approximately 30 bp shorter in patients with cancer vs. those without (15). cfDNA size distribution analysis, together with cfDNA concentration measurement, can provide prognostic information in patients with advanced cancer (16-18). Differences in cfDNA size distribution are also useful in the detection of genetic abnormalities, which can then be used to guide the choice of targeted therapy. For example, in patients with hepatocellular carcinoma, single bp resolution sequencing studies indicate that shorter plasma cfDNA fragments were more likely to include copy number alterations associated with the tumor than longer cfDNA fragments (19); however, for some patients with cancer with a low mutant allele frequency, longer cfDNA fragments have been observed (20). Therefore, cfDNA size distribution could be used as a biomarker for prognosis and for the elucidation of mechanisms of action in clinical studies.
We conducted an exploratory liquid biopsy study as part of the RELAY global, phase 3, randomized, double-blind, placebo-controlled study, which investigated the efficacy and safety of ramucirumab (RAM), a human immunoglobulin G1 vascular endothelial growth factor (VEGF) receptor 2 (VEGFR2) antagonist, plus erlotinib (ERL), an EGFR TKI, in treatment-naïve patients with EGFR-mutated metastatic NSCLC (21). A significant improvement in progression-free survival (PFS) with RAM + ERL compared with placebo (PL) plus ERL was observed {median PFS: 19.4 vs. 12.4 months; hazard ratio (HR): 0.59 [95% confidence interval (CI): 0.46–0.76; P<0.0001]}.
The objective of this RELAY exploratory, proof-of-concept, liquid biopsy addendum, specific to patients enrolled in Japan only, was to investigate (using liquid biopsy samples) whether the combination of RAM + ERL affects the occurrence of the EGFR T790M mutation and/or other mutations related to acquired EGFR TKI resistance, the association between biomarkers and treatment outcome, and the changes in cfDNA levels and fragment size. We hypothesized that cfDNA size distribution could help elucidate the mode of action of RAM in combination with an EGFR TKI in this clinical study. We present this article in accordance with the CONSORT reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-736/rc).
Methods
Study design and patients
The study design and patient population for the RELAY study have been previously described (ClinicalTrials.gov identifier: NCT02411448) (21). Briefly, RELAY was a global, phase 3, randomized, double-blind, placebo-controlled study of patients with untreated EGFR-mutated metastatic NSCLC. Two exploratory biomarker studies were conducted in RELAY; the first was conducted in the global intent-to-treat (ITT) population including Japanese patients, and the second was an optional exploratory, proof-of-concept, liquid biopsy addendum specific to RELAY patients enrolled in Japan only. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), the Council for International Organizations of Medical Sciences International Ethical Guidelines, Good Clinical Practice guidelines, and local guidelines. The protocol and addendum were approved by the ethics review boards at each site (details are provided in Table S1) and all patients provided written informed consent; patients who opted to participate in the exploratory liquid biopsy addendum provided additional consent.
Study population
Eligibility criteria for the RELAY phase 3 study have been previously published (21). Briefly, patients were ≥18 years of age (≥20 years in Japan and Taiwan) and had stage IV NSCLC, documented evidence of the EGFR exon 19 deletion (ex19del) mutation or the exon 21 L858R point mutation (ex21.L858R), ≥1 measurable lesion attributed to NSCLC [defined by Response Evaluation Criteria in Solid Tumours version 1.1 (RECIST v1.1)], Eastern Cooperative Oncology Group performance status of 0 or 1, and adequate organ function. Patients were excluded if they had received previous anti-cancer treatment for stage IIIb/IV NSCLC (except previous radiation therapy), had central nervous system metastases, or had a documented T790M EGFR mutation.
Treatment
Eligible patients were randomized (1:1) to RAM + ERL (RAM: 10 mg/kg intravenously every 2 weeks; ERL: 150 mg orally once daily) or PL + ERL (PL: intravenously every 2 weeks; ERL: 150 mg orally once daily) and were assigned using an interactive web response system. A treatment cycle was defined as 2 weeks. Patients received study treatment until disease progression, unacceptable toxicity, non-compliance, or investigator or patient decision.
Outcome measures
The primary endpoint of the RELAY phase 3 randomized portion was PFS according to RECIST v1.1. PFS was defined as the time from randomization to disease progression or death from any cause. Secondary endpoints included objective response rate (ORR), disease control rate (DCR), and duration of response (DoR), as previously described (21). Exploratory endpoints of the RELAY exploratory liquid biopsy addendum, specific to patients enrolled in Japan only, included co-occurring gene alterations at baseline detected by NGS and their potential impact on treatment outcomes (PFS, ORR, DCR, DoR) and EGFR TKI resistance mechanisms, T790M mutation rates post-study treatment discontinuation detected by droplet dPCR (ddPCR), and changes in cfDNA concentration and cfDNA fragment size throughout treatment.
Liquid biopsy sample collection and analysis
Plasma samples from patients who opted to participate in the exploratory liquid biopsy addendum study were collected for cfDNA assessment at baseline, during treatment (Cycle 4, Cycle 13, and every 6 cycles thereafter), and at 30-day post-study treatment discontinuation follow-up. Gene alterations were assessed at baseline, Cycle 4, and 30-day follow-up by NGS using the Ion AmpliSeq Colon and Lung Cancer Panel v2 (Thermo Fisher Scientific, Waltham, MA, USA). Genes included within the panels were the following: KRAS, EGFR, BRAF, PIK3CA, AKT1, ERBB2, PTEN, NRAS, STK11, MAP2K1, ALK, DDR2, CTNNB1, MET, TP53, SMAD4, FBXW7, FGFR3, NOTCH1, ERBB4, FGFR1, and FGFR2. For library preparation, cfDNA (maximum of 3,000 copies) was subjected to multiplex PCR amplification, and purified libraries were pooled and sequenced with an Ion S5 XL NGS platform and 550™ Chip Kit (Thermo Fisher Scientific, Waltham, MA, USA). Reads were aligned with the hg19 human reference genome; germline mutations were excluded with the use of the Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB) (22). Potential mutations were called using Variant Call Format version 5.12, as previously described (23). EGFR-activating mutation allele count was evaluated at all time points by ddPCR; 6 µL of cfDNA (maximum of 3,000 copies) template was added per 20 µL of ddPCR. Assays were performed in duplicate. All ddPCR assays included negative template controls and positive template controls in triplicate. Plasma cfDNA concentrations were quantified using the TaqMan RNase P Detection Reagents with the StepOne™ Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA); 1 µL of cfDNA template was subjected to real-time PCR, and cfDNA copy number was determined in reference to a standard curve. cfDNA fragment size was analyzed using the High Sensitivity DNA Chips and the 2100 Bioanalyzer Expert Software package on the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Size was determined from an external standard ladder (DNA sizing ladder), ranging from 50 up to 7,000 bp.
Statistical analysis
The analyses in this report were exploratory. The data cut-off dates were January 23, 2019 (efficacy results) and September 30, 2021 (exploratory liquid biopsy addendum results). The exploratory liquid biopsy addendum translational research (TR) population comprised patients with available baseline NGS or ddPCR results. The TR subpopulations consisted of patients with a valid baseline sample in which ≥1 gene alteration was detected by NGS (TR-NGS) or ddPCR (TR-ddPCR). Baseline gene alterations are reported for patients with any detectable gene alteration at baseline. Treatment-emergent gene alterations are reported for patients with any detectable alteration at baseline and at 30-day post-study treatment discontinuation. PFS and DoR were analyzed using the Kaplan-Meier method and compared using the unstratified log-rank test. An analysis of covariance was conducted separately for cfDNA fragment size and log-transformed cfDNA concentration. Each analysis was done by treatment arm and visit for baseline, Cycle 4, and follow-up. Post-hoc pairwise analyses with Bonferroni adjustments applied were used to compare fragment size between treatment arms within the same treatment visit and to compare across treatment visits within the treatment arm. The relationship between change in cfDNA concentration and change in cfDNA fragment size from baseline to Cycle 4 (i.e., during the early stage of study treatment) was evaluated by Pearson’s correlation coefficient for the linear relationship and Spearman’s rank correlation coefficient for the monotonic relationship.
Results
Demographic and baseline clinical characteristics
The RELAY Japanese ITT population comprised 211 patients (RAM + ERL: 106 patients; PL + ERL: 105 patients) enrolled at 41 sites (24). Of these 211 patients, 136 participated in the optional exploratory liquid biopsy addendum, which required frequent liquid biopsy sampling (Figure S1). The TR population comprised 131 patients with valid baseline (NGS and ddPCR) assay results. The TR-NGS (N=84) and TR-ddPCR (N=74) populations consisted of patients with ≥1 gene alteration detected at baseline (confirming the presence of ctDNA) by either NGS or ddPCR, respectively (Figure S1). Patients could belong to both the TR-NGS and TR-ddPCR populations; 61 patients were in both populations (EGFR ex19del: 30 patients; EGFR ex21.L858R: 31 patients). Patient and disease characteristics for the TR-NGS and TR-ddPCR populations were similar to the RELAY Japanese ITT population (Table 1). In both TR populations, most (>60%) patients were female, most (>61%) were ≥65 years of age, and >59% had never smoked. EGFR ex21.L858R mutations were identified in 40 patients in the TR-NGS group and in 42 patients in the TR-ddPCR group.
Table 1
| Variable | TR-NGS† (N=84) | TR-ddPCR† (N=74) | Japan ITT‡ (N=211) |
|---|---|---|---|
| Sex, n (%) | |||
| Female | 58 (69.0) | 48 (64.9) | 140 (66.4) |
| Age, n (%) | |||
| <65 years | 32 (38.1) | 26 (35.1) | 79 (37.4) |
| ≥65 years | 52 (61.9) | 48 (64.9) | 132 (62.6) |
| Smoking history§, n (%) | |||
| Ever | 25 (29.8) | 23 (31.1) | 62 (29.4) |
| Never | 50 (59.5) | 45 (60.8) | 127 (60.2) |
| ECOG PS 0, n (%) | 43 (51.2) | 36 (48.6) | 119 (56.4) |
| EGFR mutation¶, n (%) | |||
| Exon 19 deletion | 44 (52.4) | 32 (43.2) | 100 (47.4) |
| Exon 21 L858R mutation | 40 (47.6) | 42 (56.8) | 110 (52.1) |
†, population consists of patients from whom a valid baseline sample with ≥1 alteration detected has been obtained; ‡, previously reported in Nishio et al., 2021 (24); §, smoking history was unknown or missing for 22/211 (10%) patients in the Japan ITT population, 9/84 (11%) patients in the TR-NGS population, and 6/74 (8%) patients in the TR-ddPCR population; ¶, information on EGFR mutation was missing for 1 patient in the RAM + ERL group of the Japan ITT population. TR, translational research; NGS, next-generation sequencing; ddPCR, droplet digital polymerase chain reaction; ITT, intent-to-treat; ECOG PS, Eastern Cooperative Oncology Group performance status; EGFR, epidermal growth factor receptor; RAM, ramucirumab; ERL, erlotinib.
Efficacy
In the overall TR population, median PFS was 20.8 vs. 12.5 months [HR: 0.61 (95% CI: 0.38–0.97)] in the RAM + ERL vs. PL + ERL arms, respectively, similar to that observed in the RELAY ITT (21) and Japanese ITT (24) populations (Figure S2). Furthermore, median DoR was longer with RAM + ERL vs. PL + ERL [19.0 vs. 11.1 months; HR: 0.51 (95% CI: 0.31–0.84)] (Table 2) and was similar to that observed in the Japanese ITT population (24).
Table 2
| Response | TR population | ||
|---|---|---|---|
| RAM + ERL (N=70) | PL + ERL (N=61) | Overall (N=131) | |
| CR | 1 (1.4) | 0 | 1 (0.8) |
| PR | 53 (75.7) | 44 (72.1) | 97 (74.1) |
| SD | 12 (17.1) | 15 (24.6) | 27 (20.6) |
| Progressive disease | 1 (1.4) | 1 (1.6) | 2 (1.5) |
| Not assessed | 3 (4.3) | 1 (1.6) | 4 (3.1) |
| ORR (CR + PR) | 54 (77.1) | 44 (72.1) | 98 (74.8) |
| DCR (CR + PR + SD) | 66 (94.3) | 59 (96.7) | 125 (95.4) |
| Duration of response† | |||
| Events | 29 (53.7) | 33 (75.0) | |
| Median (95% CI) (months) | 19.0 (15.0–NA) | 11.1 (9.0–16.5) | |
| HR (95% CI) | 0.51 (0.31–0.84) | ||
Data are n (%) except where indicated. †, in patients who responded (RAM + ERL: n=54; PL + ERL: n=44). TR, translational research; RAM, ramucirumab; ERL, erlotinib; PL, placebo; CR, complete response; PR, partial response; SD, stable disease; ORR, objective response rate; DCR, disease control rate; CI, confidence interval; NA, not available; HR, hazard ratio.
Co-occurring gene alterations
Co-occurring baseline gene alterations detected by NGS in ≥5 patients in the TR-NGS population were TP53 (36/84 patients, 42.9%), PTEN (6/84 patients, 7.1%), and KRAS (5/84 patients, 6.0%) (Table 3). Comparison of PFS by baseline TP53 status and treatment arm did not establish a prognostic or predictive relationship (data not shown).
Table 3
| Gene | RAM + ERL (N=41), n (%) | PL + ERL (N=43), n (%) | Overall (N=84), n (%) |
|---|---|---|---|
| TP53 | 18 (43.9) | 18 (41.9) | 36 (42.9) |
| PTEN | 1 (2.4) | 5 (11.6) | 6 (7.1) |
| KRAS | 3 (7.3) | 2 (4.7) | 5 (6.0) |
| Other EGFR† | 2 (4.9) | 2 (4.7) | 4 (4.8) |
| T790M | 1 (2.4) | 0 | 1 (1.2) |
| CTNNB1 | 0 | 3 (7.0) | 3 (3.6) |
| MET | 2 (4.9) | 0 | 2 (2.4) |
| BRAF | 1 (2.4) | 0 | 1 (1.2) |
| FBXW7 | 0 | 1 (2.3) | 1 (1.2) |
| FGFR3 | 1 (2.4) | 0 | 1 (1.2) |
| PIK3CA | 0 | 1 (2.3) | 1 (1.2) |
| SMAD4 | 0 | 1 (2.3) | 1 (1.2) |
†, EGFR mutation excluding EGFR‑activating mutations exon 19 deletion and exon 21 L858R mutation. NGS, next-generation sequencing; TR, translational research; RAM, ramucirumab; ERL, erlotinib; PL, placebo; EGFR, epidermal growth factor receptor.
Treatment-emergent gene alterations detected by NGS at post-study treatment discontinuation included EGFR, FGFR3, KRAS, and TP53 (Table 4). Treatment-emergent EGFR mutations included T790M (by NGS and ddPCR) and H870R (assessed by NGS only) (Table 4). T790M mutation rates by ddPCR were 54.5% (6/11 patients) in the RAM + ERL arm and 41.2% (7/17 patients) in the PL + ERL arm; T790M mutation rates by NGS were 22.2% (2/9 patients) in the RAM + ERL arm and 29.4% (5/17 patients) in the PL + ERL arm.
Table 4
| Gene alterations | RAM + ERL, n (%) | PL + ERL, n (%) | Overall, n (%) |
|---|---|---|---|
| Mutations detected by NGS† | |||
| EGFR‡ | 2 (22.2) | 5 (29.4) | 7 (26.9) |
| H870R | 1 (11.1) | 0 | 1 (3.8) |
| T790M | 2 (22.2) | 5 (29.4) | 7 (26.9) |
| FGFR3 | 1 (11.1) | 0 | 1 (3.8) |
| KRAS | 2 (22.2) | 0 | 2 (7.7) |
| TP53 | 3 (33.3) | 1 (5.9) | 4 (15.4) |
| None | 4 (44.4) | 11 (64.7) | 15 (57.7) |
| Mutations detected by ddPCR§ | |||
| T790M | 6 (54.5) | 7 (41.2) | 13 (46.4) |
†, RAM + ERL, N=9; PL + ERL, N=17; Overall, N=26; ‡, one patient had 2 treatment-emergent EGFR mutations (T790M and H870R); 3 patients did not have EGFR‑activating mutations detected in ctDNA at baseline but did have these mutations detected at 30-day post-study treatment discontinuation, consistent with their local baseline testing (2 exon 19 deletion, 1 L858R), and were not included in this treatment-emergent summary; §, RAM + ERL, N=11; PL + ERL, N=17; Overall, N=28. RAM, ramucirumab; ERL, erlotinib; PL, placebo; NGS, next-generation sequencing; ddPCR, droplet digital polymerase chain reaction; EGFR, epidermal growth factor receptor; ctDNA, circulating tumor DNA.
EGFR-activating mutant alleles and treatment outcome
When assessed according to dichotomized baseline EGFR-activating mutation allele count in this data set (low or high allele count was below or above the baseline median mutation allele count of 102, respectively), no difference in PFS was observed in either treatment arm (Figure 1A). The number of EGFR-activating mutation alleles decreased from baseline at Cycle 4 and was sustained throughout treatment in both treatment arms; at follow-up, allele count increased in the PL + ERL arm but not in the RAM + ERL arm (Figure S3). At baseline, an EGFR-activating mutation was detectable in 94.6% (35/37) of patients in the RAM + ERL arm and 94.6% (35/37) of patients in the PL + ERL arm. At Cycle 4, no EGFR-activating mutation was detectable in 78% (21/27) of patients in the RAM + ERL arm and 67% (20/30) of patients in the PL + ERL arm. PFS was improved for patients with no detectable EGFR-activating mutation at Cycle 4 (median PFS: RAM + ERL: not reached vs. PL + ERL: 12.52 months; HR: 0.28, 95% CI: 0.09–0.73) vs. those with detectable EGFR-activating mutation (median PFS: RAM + ERL, 12.65 months vs. PL + ERL, 9.64 months; HR: 0.66, 95% CI: 0.14–2.28) (Figure 1B).

Total cfDNA concentration and fragment size
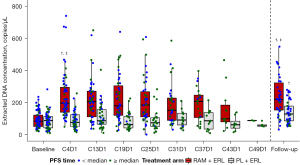
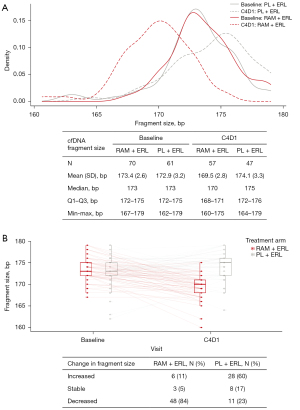
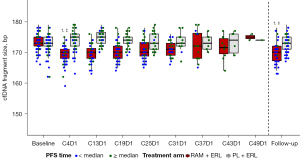
Total cfDNA concentration increased in the RAM+ERL arm from Cycle 4 and was sustained throughout treatment [mean (standard deviation): baseline, 92.1 (52.58) vs. Cycle 4, 239.4 (144.96) copies/µL, P for log-transformed data <0.0001; baseline vs. follow-up: 314.6 (512.38) copies/µL, P for log-transformed data <0.0001] (Figure 2). cfDNA fragment size was similar at baseline in the 2 treatment arms [mean (standard deviation): RAM + ERL, 173.4 (2.6) vs. PL + ERL, 172.9 (3.2) bp; P=0.35] but was shorter in the RAM + ERL arm compared with the PL + ERL arm at Cycle 4, respectively [mean (standard deviation): 169.5 (2.8) vs. 174.1 (3.3) bp; P<0.0001] (Figure 3A). No change in pattern for subgroups by EGFR-activating mutation (detected/not detected) at baseline was observed (data not shown). cfDNA fragment size decreased (baseline vs. Cycle 4) in 84% (RAM + ERL; 48/57) and 23% (PL + ERL; 11/47) of paired patient samples (Figure 3B). A trend for shorter cfDNA fragment size in the RAM + ERL arm compared with the PL + ERL arm was observed throughout treatment and at follow-up (Figure 4). A negative correlation between change in cfDNA concentration vs. change in cfDNA fragment size from baseline to Cycle 4 was identified in the overall TR population (RAM + ERL and PL + ERL treatment arms combined) (Figure 5).

Discussion
This exploratory liquid biopsy addendum of the RELAY phase 3 study of RAM + ERL vs. PL + ERL examined gene alterations, total cfDNA concentration, and cfDNA fragment size in patient-derived liquid biopsy samples throughout and after treatment. Improvement in PFS was observed for patients with no detectable EGFR-activating mutation at Cycle 4 in liquid biopsy samples compared with those with a detectable EGFR-activating mutation. Furthermore, throughout treatment, increased levels of total cfDNA were detected in the RAM + ERL patient samples but were not apparent in the PL + ERL patient samples, suggesting an enhanced anti-tumor effect with the addition of RAM to ERL. Fragment size of the total cfDNA content in the RAM + ERL samples was shorter than that in the PL + ERL samples, suggesting that the increased total cfDNA levels in the RAM + ERL arm are likely due to increased tumor cell apoptosis. These results provide insight into possible mechanisms of resistance and/or efficacy of RAM in addition to ERL in the treatment of NSCLC.
EGFR mutation-positive tumors often contain co-occurring gene alterations, the identification of which will vary depending on the detection method used (25). Common co-occurring gene alterations include TP53, PI3KCA, RB1, or CTNNB1 and will vary in frequency in early-stage vs. advanced-stage tumors (26). TP53, PI3KCA, and RB1 co-occurring alterations have a prognostic impact on worse clinical outcomes in EGFR mutation-positive NSCLC treated with EGFR TKI therapy (26-29). In this study of Japanese patients with EGFR mutation-positive NSCLC, common baseline gene alterations co-occurring with an EGFR-activating mutation in ≥5 patients included TP53, PTEN, and KRAS. Treatment-emergent gene alterations included EGFR T790M and TP53 in both treatment arms, and EGFR H870R, FGFR3, and KRAS in the RAM + ERL arm. One patient harbored the previously described EGFR H870R mutation that in combination with ex21.L858R may lead to resistance to the EGFR TKI gefitinib (30,31).
Of patients receiving first- or second-generation EGFR TKI therapy for the treatment of EGFR mutation-positive NSCLC, 50–60% will acquire the EGFR T790M resistance mutation (32), after which the only effective EGFR TKI therapy available is a third-generation EGFR TKI (2,3). High sensitivity and quantitative concordance of amplicon-based plasma NGS compared with ddPCR in detecting resistance mechanisms, such as T790M, have been demonstrated (33). In the RELAY global ITT and Japanese ITT populations, post-progression T790M rates detected by Guardant 360 NGS were similar for RAM + ERL vs. PL + ERL (21,24). In this exploratory liquid biopsy study, T790M rates detected by ddPCR and NGS at post-study treatment discontinuation differed, which may have been due to the different sensitivity of the detection methods. Regardless of the testing method used, the observed cumulative frequency of the T790M mutation detected in liquid biopsy samples was not affected by the addition of RAM to ERL. These results suggest that first-line RAM + ERL could provide the opportunity for second-line molecular targeted therapy by third-generation EGFR TKI treatment that is active against the T790M mutation.
EGFR-activating mutation allele count/allele frequency in liquid biopsy samples could be a potential biomarker for response to treatment. Buder et al. (34) showed that patients with advanced EGFR T790M-mutated NSCLC who responded to second-line osimertinib had a significantly lower EGFR-activating mutation allele frequency at baseline than patients who did not respond, and that a higher allele frequency in plasma ctDNA was associated with shorter PFS. In our study, no difference in PFS was observed between high and low baseline count of EGFR-activating mutation alleles with RAM + ERL or PL + ERL treatment. However, PFS was improved for patients with no detectable EGFR-activating mutation alleles at Cycle 4 compared with those who did have detectable EGFR-activating mutation alleles. These results suggest that monitoring of circulating EGFR-activating mutation alleles at Cycle 4 could provide insights regarding likely clinical benefit. Although the count of EGFR-activating mutation alleles at baseline was slightly different between treatment arms, it was suppressed to a similar level throughout treatment and was only sustained at post-study treatment discontinuation follow-up with RAM + ERL treatment, suggesting that RAM enhances the sustained anti-tumor effect of ERL on EGFR-mutated cells. From the viewpoint of molecular residual disease detection, monitoring the EGFR-activating mutation fraction by liquid biopsy may predict disease progression; however, further investigation is required for the clinical application of such monitoring.
The total cfDNA content is the sum of tumor-derived ctDNA, such as ctDNA from ex19del- or ex21.L858R-mutated tumor cells and other tumor cells, cfDNA from normal cells in the tumor microenvironment (e.g., stroma and pericytes), and cfDNA from primary hematopoietic origin, with ctDNA fragment lengths frequently being shorter than normal cfDNA fragments (35). Moreover, ctDNA and cfDNA are considered to be real-time snapshots of the tumor microenvironment due to their short half-lives. However, we need to interpret cfDNA profiling carefully because there are 2 different mechanisms by which cfDNA is released into the bloodstream; one is active release of cfDNA from the tumor and the other is passive release from dying cells by treatment (36,37). In our analysis, despite the number of EGFR-activating mutation alleles being suppressed at Cycle 4 and throughout treatment in both treatment arms, increased levels of total cfDNA were detected throughout treatment in the RAM+ERL arm but not in the PL + ERL arm. Furthermore, fragment size of the total cfDNA content in the RAM + ERL arm was shorter than at baseline and then in the PL + ERL arm throughout treatment, indicating that the increased levels of total cfDNA in the RAM + ERL arm were likely due to increased tumor cell apoptosis of cells other than EGFR mutation-positive cells. This phenomenon suggests that RAM may promote continuous tumor cell apoptosis throughout treatment with RAM + ERL (Figure S4). However, further investigation is required for the clinical application of monitoring the size of cfDNA.
Although longitudinal evaluation of mutations was pre-specified, this analysis was exploratory and intended to be hypothesis generating. Nevertheless, our results suggest that RAM enhances the sustained anti-tumor effect of ERL on EGFR mutation-positive tumors. We hypothesize 2 scientific reasons behind this result. The first is a direct effect on the tumor by dual blockade of EGFR and VEGF. Tumor cells express VEGFR2 (38), and the EGFR and VEGF pathways are interconnected (39). Furthermore, VEGFR2 inhibition also has a direct anti-tumor effect on cancer cells. In xenograft mouse models of EGFR-, ALK-, or ROS1-altered NSCLC, the combination of VEGFR2 blockade with molecular targeted agents showed enhanced anti-tumor effects of the molecular targeted agents (40). Thus, blockade of VEGFR2 with RAM may similarly enhance the anti-tumor effect of EGFR blockade by ERL. The second scientific reason that we hypothesize for the enhanced anti-tumor effect of ERL on EGFR-mutated cells by RAM is an indirect effect on the tumor by inhibition of angiogenesis by RAM. Inhibition of angiogenesis normalizes tumor vessels (41,42), and the improved vasculature enhances drug (ERL) delivery to the tumor (43,44), thereby increasing its efficacy. ERL increases the apoptosis of EGFR mutation-positive tumor cells (45), thus reducing the count of EGFR-activating mutation alleles.
In this study, we observed that the count of EGFR-activating mutation alleles was suppressed throughout treatment in both treatment arms (ERL effect). However, despite this suppression, most patients will progress, indicating that the tumor eventually becomes treatment resistant. One reason for this may be intra-tumoral heterogeneity arising from genetic and epigenetic alterations derived from genomic and chromosomal instability and different patterns of clonal evolution; such heterogeneity has been reported in NSCLC (46,47). Furthermore, in this study, we observed short fragment-sized cfDNA increased in the RAM + ERL arm but not the PL + ERL arm. We hypothesize that the increased concentration of short fragment-sized cfDNA observed with RAM + ERL was derived from tumor cells other than EGFR mutation-positive cells; this is supported by observations in a previous study, whereby inhibition of angiogenesis with an anti-VEGFR2 antibody (DC101) led to an increase in apoptosis of endothelial cells followed by apoptosis of tumor cells (48,49), which would result in a release of fragmented DNA. Indeed, RAM monotherapy has been shown to have survival benefits in patients with advanced gastric or gastroesophageal junction adenocarcinoma progressing after first-line chemotherapy, presumedly by targeting the VEGF pathway and inhibiting angiogenesis (50). This additional anti-tumor effect of RAM by inhibiting VEGFR2 on both tumor and non-tumor cells may contribute to the PFS benefit observed in the RELAY study with RAM + ERL compared with PL + ERL.
This study used both ddPCR and NGS methodologies to identify potential biomarkers in liquid biopsy samples, which, for most analyses, were taken at multiple time points throughout treatment and at post-study treatment discontinuation follow-up. NGS or an electrophoretic mobility assay such as the Bioanalyzer can be used to determine the proportion of ctDNA in a sample (51). In this exploratory study, the Bioanalyzer was used to compare the size distribution of cfDNA. The Bioanalyzer has the advantage of high sensitivity (down to 5 pg/µL for fragment analysis) and requires a smaller sample volume (1 µL for nucleic acids). However, the study was limited to patients with both baseline and 30-day follow-up samples, thus limiting the sample size of the study and making it difficult to draw inferences regarding treatment-emergent mutations. Furthermore, the current data may be biased toward patients who discontinued study treatment, some of whom were early progressors, because patients who were still on treatment were not included.
Conclusions
In conclusion, this biomarker analysis indicated that the count of EGFR-activating mutation alleles was suppressed, total cfDNA concentration was increased, and short fragment-sized cfDNA increased with RAM+ERL. Taken together, these results suggest that the additional anti-tumor effect of RAM by inhibiting VEGFR2 on tumor and non-tumor cells may contribute to the PFS benefit observed in the RELAY study with RAM + ERL compared with PL + ERL.
Acknowledgments
We thank all participants, their caregivers, investigators, and study site staff who were involved in the study.
Funding: This work was supported by Eli Lilly and Company, manufacturer/licensee of ramucirumab. The funder was involved in the study design, data collection, data analysis, and preparation of the manuscript. Medical writing assistance was provided by Prudence Stanford, PhD, CMPP, and Rebecca Lew, PhD, CMPP, of ProScribe – Envision Pharma Group, and was funded by Eli Lilly Japan K.K. ProScribe’s services complied with international guidelines for Good Publication Practice.
Footnote
Reporting Checklist: The authors have completed the CONSORT reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-736/rc
Trial Protocol: Available in the Supplementary Appendix of Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20(12):1655-69. doi:10.1016/S1470-2045(19)30634-5.
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-736/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-736/coif). The authors report the funding from Eli Lilly and Company. Kazuto Nishio reports grants or contracts from Eli Lilly Japan, Clinical Research Support Center Kyushu, Hitachi, Nichirei Biosciences, Nippon Boehringer Ingelheim, North East Japan Study Group, Osakaminami Hospital, Sysmex Corporation, Thoracic Oncology Research Group, and West Japan Oncology Group; consulting fees from Eli Lilly Japan, Otsuka Pharmaceutical, Solasia Pharma, and SymBio Pharmaceuticals; and honoraria for lectures from Amgen, AstraZeneca, Boehringer Ingelheim Japan, Bristol Myers Squibb, Chugai Pharmaceutical, Eisai, Eli Lilly Japan, Fujirebio, Guardant Health, Janssen, Merck Biopharma, Merck Sharp & Dohme, Novartis, Ono Pharmaceutical, Pfizer, Roche Diagnostics, Sanofi, Takeda, and Yakult Honsha. Kazuko Sakai declares support for the present manuscript by Eli Lilly and Company. MN has received payment or honoraria from Astellas, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Chugai Pharmaceutical, Eli Lilly, Merck Serono, Merck Sharp & Dohme, Novartis, Ono Pharmaceutical, Pfizer, Sankyo Healthcare, and Taiho Pharmaceutical. TS reports grants or contracts to their institution from AbbVie, Chugai Pharmaceutical, Daiichi Sankyo, Eli Lilly Japan, Kissei Pharmaceutical, Merck Sharp & Dohme, Novartis, Pfizer Japan, and Takeda Pharmaceutical; honoraria for lectures from AstraZeneca, Bristol Myers Squibb, Chugai Pharmaceutical, Covidien Japan, Daiichi Sankyo, Eli Lilly Japan, Kyowa Hakko Kirin, Merck Sharp & Dohme, Mochida Pharmaceutical, Nippon Boehringer Ingelheim, Novartis, Ono Pharmaceutical, Pfizer Japan, Taiho Pharmaceutical, Takeda, and Towa Pharmaceutical; and employment with Precision Medicine Asia. CVG and MC are employees and minor shareholders of Eli Lilly and Company. TM and SE are employees of Eli Lilly Japan K.K. and are minor shareholders of Eli Lilly and Company. Kazuhiko Nakagawa reports grants or contracts to their institution from AbbVie, Amgen, AstraZeneca K.K., Bayer Yakuhin, Bristol Myers Squibb, Chugai Pharmaceutical, Covance Japan, Daiichi Sankyo, Eisai, Eli Lilly Japan K.K., EP-CRSU, EPS Corporation, EPS International, GlaxoSmithKline K.K., IQVIA Services Japan K.K., Janssen Pharmaceutical K.K., Japan Clinical Research Operations, Kissei Pharmaceutical, Mebix, Medical Research Support, Merck Biopharma, Merck Sharp & Dohme K.K., Mochida Pharmaceutical, Nippon Boehringer Ingelheim, Nippon Kayaku, Novartis Pharma K.K., Ono Pharmaceutical, Otsuka Pharmaceutical, Parexel, Pfizer Japan, Pfizer R&D Japan G.K., PPD-SNBL K.K., PRA Health Sciences, Sanofi K.K., SRL, SymBio Pharmaceuticals, Syneos Health Clinical K.K., Sysmex Corporation, Taiho Pharmaceutical, and Takeda; consulting fees from Eli Lilly Japan K.K., KYORIN Pharmaceutical, Ono Pharmaceutical, and Pfizer Japan; payment or honoraria from 3H Clinical Trial, AbbVie, Amgen, AstraZeneca K.K., Bayer Yakuhin, Bristol Myers Squibb K.K., Care Net, Chugai Pharmaceutical, CMIC, CMIC ShiftZero K.K., Eli Lilly Japan K.K., Japan Clinical Research Operations, KYORIN Pharmaceutical, Kyowa Kirin, Life Technologies Japan, Medical Mobile Communications, Medical Review, Merck Serono, Merck Sharp & Dohme K.K., Neo Communications, Nikkei Business Publications, Nippon Boehringer Ingelheim, Nippon Kayaku, Novartis Pharma K.K., Ono Pharmaceutical, Pfizer Japan, Roche Diagnostics K.K., Taiho Pharmaceutical, TAIYO Pharma, Takeda Pharmaceutical, and YODOSHA; and patents planned, issued, or pending with Daiichi Sankyo and their institution. The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chevallier M, Borgeaud M, Addeo A, et al. Oncogenic driver mutations in non-small cell lung cancer: Past, present and future. World J Clin Oncol 2021;12:217-37. [Crossref] [PubMed]
- Wu YL, Planchard D, Lu S, et al. Pan-Asian adapted Clinical Practice Guidelines for the management of patients with metastatic non-small-cell lung cancer: a CSCO-ESMO initiative endorsed by JSMO, KSMO, MOS, SSO and TOS. Ann Oncol 2019;30:171-210. [Crossref] [PubMed]
- Akamatsu H, Ninomiya K, Kenmotsu H, et al. The Japanese Lung Cancer Society Guideline for non-small cell lung cancer, stage IV. Int J Clin Oncol 2019;24:731-70. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Guibert N, Pradines A, Favre G, et al. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur Respir Rev 2020;29:190052. [Crossref] [PubMed]
- Wu Z, Yang Z, Dai Y, et al. Update on liquid biopsy in clinical management of non-small cell lung cancer. Onco Targets Ther 2019;12:5097-109. [Crossref] [PubMed]
- Ishii H, Azuma K, Sakai K, et al. Digital PCR analysis of plasma cell-free DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: Correlation with paired tumor samples. Oncotarget 2015;6:30850-8. [Crossref] [PubMed]
- Xu S, Lou F, Wu Y, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett 2016;370:324-31. [Crossref] [PubMed]
- Parsons HA, Rhoades J, Reed SC, et al. Sensitive Detection of Minimal Residual Disease in Patients Treated for Early-Stage Breast Cancer. Clin Cancer Res 2020;26:2556-64. [Crossref] [PubMed]
- Zheng YW, Chan KC, Sun H, et al. Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: a transplantation model. Clin Chem 2012;58:549-58. [Crossref] [PubMed]
- Ivanov M, Baranova A, Butler T, et al. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics 2015;16:S1. [Crossref] [PubMed]
- Straver R, Oudejans CB, Sistermans EA, et al. Calculating the fetal fraction for noninvasive prenatal testing based on genome-wide nucleosome profiles. Prenat Diagn 2016;36:614-21. [Crossref] [PubMed]
- Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10:eaat4921. [Crossref] [PubMed]
- Underhill HR. Leveraging the Fragment Length of Circulating Tumour DNA to Improve Molecular Profiling of Solid Tumour Malignancies with Next-Generation Sequencing: A Pathway to Advanced Non-invasive Diagnostics in Precision Oncology? Mol Diagn Ther 2021;25:389-408. [Crossref] [PubMed]
- Chen E, Cario CL, Leong L, et al. Cell-free DNA concentration and fragment size as a biomarker for prostate cancer. Sci Rep 2021;11:5040. [Crossref] [PubMed]
- Lapin M, Oltedal S, Tjensvoll K, et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J Transl Med 2018;16:300. [Crossref] [PubMed]
- Yamamoto Y, Uemura M, Nakano K, et al. Increased level and fragmentation of plasma circulating cell-free DNA are diagnostic and prognostic markers for renal cell carcinoma. Oncotarget 2018;9:20467-75. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]
- Liu X, Lang J, Li S, et al. Fragment Enrichment of Circulating Tumor DNA With Low-Frequency Mutations. Front Genet 2020;11:147. [Crossref] [PubMed]
- Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:1655-69. [Crossref] [PubMed]
- Narahara M, Higasa K, Nakamura S, et al. Large-scale East-Asian eQTL mapping reveals novel candidate genes for LD mapping and the genomic landscape of transcriptional effects of sequence variants. PLoS One 2014;9:e100924. [Crossref] [PubMed]
- Ohira T, Sakai K, Matsubayashi J, et al. Tumor volume determines the feasibility of cell-free DNA sequencing for mutation detection in non-small cell lung cancer. Cancer Sci 2016;107:1660-6. [Crossref] [PubMed]
- Nishio K, Seto T, Nishio M, et al. Ramucirumab Plus Erlotinib Versus Placebo Plus Erlotinib in Patients With Untreated Metastatic EGFR-Mutated NSCLC: RELAY Japanese Subset. JTO Clin Res Rep 2021;2:100171. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. J Clin Med 2020;9:1047. [Crossref] [PubMed]
- Qiu X, Wang Y, Liu F, et al. Survival and prognosis analyses of concurrent PIK3CA mutations in EGFR mutant non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors. Am J Cancer Res 2021;11:3189-200.
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- De Pas T, Toffalorio F, Manzotti M, et al. Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations. J Thorac Oncol 2011;6:1895-901. [Crossref] [PubMed]
- Tam IY, Leung EL, Tin VP, et al. Double EGFR mutants containing rare EGFR mutant types show reduced in vitro response to gefitinib compared with common activating missense mutations. Mol Cancer Ther 2009;8:2142-51. [Crossref] [PubMed]
- Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. [Crossref] [PubMed]
- Ding PN, Becker T, Bray V, et al. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thorac Cancer 2019;10:1879-84. [Crossref] [PubMed]
- Buder A, Hochmair MJ, Filipits M. The Allele Frequency of EGFR Mutations Predicts Survival in Advanced EGFR T790M-Positive Non-small Cell Lung Cancer Patients Treated with Osimertinib. Target Oncol 2021;16:77-84. [Crossref] [PubMed]
- Underhill HR, Kitzman JO, Hellwig S, et al. Fragment Length of Circulating Tumor DNA. PLoS Genet 2016;12:e1006162. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Lyu X, Tsui YM, Ho DW, et al. Liquid Biopsy Using Cell-Free or Circulating Tumor DNA in the Management of Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol 2022;13:1611-24. [Crossref] [PubMed]
- Seto T, Higashiyama M, Funai H, et al. Prognostic value of expression of vascular endothelial growth factor and its flt-1 and KDR receptors in stage I non-small-cell lung cancer. Lung Cancer 2006;53:91-6. [Crossref] [PubMed]
- Le X, Nilsson M, Goldman J, et al. Dual EGFR-VEGF Pathway Inhibition: A Promising Strategy for Patients With EGFR-Mutant NSCLC. J Thorac Oncol 2021;16:205-15. [Crossref] [PubMed]
- Watanabe H, Ichihara E, Kayatani H, et al. VEGFR2 blockade augments the effects of tyrosine kinase inhibitors by inhibiting angiogenesis and oncogenic signaling in oncogene-driven non-small-cell lung cancers. Cancer Sci 2021;112:1853-64. [Crossref] [PubMed]
- Tong RT, Boucher Y, Kozin SV, et al. Vascular normalization by vascular endothelial growth factor receptor 2 blockade induces a pressure gradient across the vasculature and improves drug penetration in tumors. Cancer Res 2004;64:3731-6. [Crossref] [PubMed]
- Tsukada Y, Muramatsu F, Hayashi Y, et al. An in vivo model allowing continuous observation of human vascular formation in the same animal over time. Sci Rep 2021;11:745. [Crossref] [PubMed]
- Chauhan VP, Stylianopoulos T, Martin JD, et al. Normalization of tumour blood vessels improves the delivery of nanomedicines in a size-dependent manner. Nat Nanotechnol 2012;7:383-8. [Crossref] [PubMed]
- Chatterjee S, Wieczorek C, Schöttle J, et al. Transient antiangiogenic treatment improves delivery of cytotoxic compounds and therapeutic outcome in lung cancer. Cancer Res 2014;74:2816-24. [Crossref] [PubMed]
- Amann J, Kalyankrishna S, Massion PP, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res 2005;65:226-35.
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Passaro A, Malapelle U, Del Re M, et al. Understanding EGFR heterogeneity in lung cancer. ESMO Open 2020;5:e000919. [Crossref] [PubMed]
- Bruns CJ, Liu W, Davis DW, et al. Vascular endothelial growth factor is an in vivo survival factor for tumor endothelium in a murine model of colorectal carcinoma liver metastases. Cancer 2000;89:488-99.
- Sweeney P, Karashima T, Kim SJ, et al. Anti-vascular endothelial growth factor receptor 2 antibody reduces tumorigenicity and metastasis in orthotopic prostate cancer xenografts via induction of endothelial cell apoptosis and reduction of endothelial cell matrix metalloproteinase type 9 production. Clin Cancer Res 2002;8:2714-24.
- Fuchs CS, Tomasek J, Yong CJ, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014;383:31-9. [Crossref] [PubMed]
- Alcaide M, Cheung M, Hillman J, et al. Evaluating the quantity, quality and size distribution of cell-free DNA by multiplex droplet digital PCR. Sci Rep 2020;10:12564. [Crossref] [PubMed]


