
KMT2C expression and DNA homologous recombination repair factors in lung cancers with a high-grade fetal adenocarcinoma component
Highlight box
Key findings
• High-grade fetal adenocarcinoma (H-FLAC) components showed lower expression of KMT2C and homologous recombination repair (HRR) factors than common lung adenocarcinomas.
• KMT2C expression was positively correlated with the expression of HRR factors.
What is known and what is new?
• Only two studies using cancer cell lines previously reported that KMT2C directly and/or indirectly promotes the expression of HRR factors, and KMT2C dysfunction was associated with sensitivity to PARP inhibitors.
• KMT2C expression was significantly lower in surgically resected H-FLACs than in that of common adenocarcinomas.
What is the implication, and what should change now?
• H-FLACs with a high degree of KMT2C dysfunction may be highly sensitive to PARP inhibitors. PARP inhibitors should only be considered for treatment of H-FLAC after further studies.
Introduction
Background
Fetal adenocarcinoma of the lung is a rare variant of lung adenocarcinoma, displaying characteristic histology resembling the fetal lung epithelium of the pseudoglandular stage, and is classified into low-grade and high-grade forms (1,2). Low-grade fetal lung adenocarcinoma (L-FLAC) frequently occurs in young- to middle-aged women with a smoking history, and its histology often shows morule formation (1,3). High-grade fetal adenocarcinoma (H-FLAC) predominantly occurs in elderly men with a history of heavy smoking and histologically shows marked nuclear atypia and no morule formation (1,4-7). H-FLAC usually coexists with other components such as common adenocarcinoma, enteric adenocarcinoma, and neuroendocrine carcinoma (4,6,7). Similar to micropapillary adenocarcinomas, lung adenocarcinomas with an H-FLAC component have poor prognoses (7). L-FLAC recurrently exhibits mutations of CTNNB1 and DICER1, which are rare in H-FLACs (8-10). Therefore, L- and H-FLACs have diverse clinicopathological and molecular features and may develop through distinct tumorigenic pathways (1,6,11).
Our recent study (11) showed that lung cancers with H-FLAC components rarely harboured druggable driver gene mutations but exhibited a high frequency of KMT2C mutations (6/16 cases, 38%). KMT2C encodes a histone H3K4 monomethyltransferase, and its expression leads to enhancer activation (12). Although little is known about the consequences of KMT2C dysfunction in cancer cells, Rampias et al. recently reported that low KMT2C activity in bladder cancer cells is associated with a decrease in ATM, ATR, BRCA1, and BRCA2 expression, resulting in the impairment of homologous recombination-mediated DNA double-strand break repair and high sensitivity to olaparib, a poly (ADP-ribose) polymerase (PARP1/2) inhibitor (13). PARP1 and PARP2 are enzymes involved in the DNA damage response and facilitate DNA single-strand break repair (14). PARP1/2 inhibitors lead to genomic instability and cell death in tumour cells with mutations in BRCA1/2 genes, which encode proteins involved in key stages of homologous recombination in DNA double-strand break repair (15).
In the past few decades, platinum-based cytotoxic chemotherapy has been the systemic treatment of choice for unresectable advanced lung cancers. Currently, targeted molecular therapy for lung cancers is rapidly developing, and various kinase inhibitors are used clinically to treat non-small-cell lung cancers with targetable driver mutations of EGFR, ALK, RET, ROS1, BRAF, and MET genes (16). First-line immunotherapy with a PD-L1 checkpoint inhibitor also represents a marked development in the treatment of advanced non-small-cell lung cancers with high PD-L1 levels and no driver mutations (17). The lack of targetable driver mutations causes most cases of H-FLACs to be ineligible for current targeted molecular therapy with kinase inhibitors. Additionally, H-FLACs tend to have no/low PD-L1 expression; therefore, immunotherapy may also have little effect on H-FLAC cases (11).
Rationale and knowledge gap
As H-FLACs harbour a high frequency of KMT2C mutations, epigenetic aberrations due to KMT2C dysfunction seemed to be associated with the biological features of H-FLACs. KMT2C dysfunction in H-FLACs may lead to impaired DNA damage repair through homologous recombination and PARP1/2 inhibitor sensitization. However, there are few studies on the expression and functional consequences of KMT2C in non-small cell lung cancer tissues. To our knowledge, the effect of KMT2C aberration on H-FLACs has not been investigated yet.
Objective
To investigate the functional consequences of KMT2C expression in H-FLAC, we first obtained an RNA expression profile by RNA sequencing and identified differentially expressed genes and pathways between H-FLAC and common primary lung adenocarcinomas. Next, we focused on homologous recombination repair (HRR) factors, such as ATM, ATR, BRCA1, and BRCA2, and performed immunohistochemical analysis of HRR factors to evaluate the levels of protein expression in H-FLAC and common adenocarcinomas and then examined the association between KMT2C and HRR factor expression in H-FLAC. Additionally, we focused on RAD51 which is another critical effector of HRR factor (18) and investigated RAD51 expression in H-FLAC. We present this article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-137/rc).
Methods
Case selection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Kanagawa Cancer Center (approval No. H28-Res60) in Yokohama, Japan, and individual consent for this retrospective analysis was waived. We obtained 16 cases (F1–F16) of surgically resected primary lung cancers with an H-FLAC components present in various proportions (at least 5% of the total tumour volume). All cases were previously reported (11). The case numbers of lung cancers with an H-FLAC component were the same as those in the previous report, and KMT2C mutations were detected in F4, F5, F8, F10, F12, and F13 in the previous study (11). H-FLAC was histologically defined as previously described (7). Additionally, for comparative analyses, we collected common primary lung adenocarcinomas, which were surgically resected at the Kanagawa Cancer Center, and randomly selected 16 cases (C1–C16) that were matched with the H-FLAC cases by clinicopathological factors such as age, sex, smoking status, pathological stage, and year of operation. The common adenocarcinoma cases selected consisted of non-mucinous neoplastic glands with a complex admixture of histologic patterns (lepidic, acinar, papillary, micropapillary, and solid) and lacked components of special variants, including invasive mucinous, colloid, fetal, and enteric-type adenocarcinomas. All resected tissues were fixed in 10% formalin, embedded in paraffin, and stained with haematoxylin and eosin (HE). Histological assessment and classification were performed following the World Health Organization classification of lung tumours (2). Although the Union for International Cancer Control (UICC) published the 8th edition of the UICC TNM classification of malignant tumours in 2017 (19), the lung cancer stages in this study were evaluated using the 7th edition (20), owing to the difficulty in the accurate retrospective measurement of invasive sizes of past lung cancer cases.
RNA sequencing
Based on the area of tumour tissues in the HE-stained section, total RNA was isolated from formalin-fixed paraffin-embedded tumour tissues of each lung cancer case (F1–16 and C1–16) and subjected to transcriptome analysis by RNA sequencing. In the H-FLAC cases (F1–16), tissues from only the H-FLAC components were manually obtained through macrodissection and used for the analysis. Total RNA was extracted using a NucleoSpin totalRNA FFPE XS kit (Macherey-Nagel GmbH & Co. KG, Germany), according to the manufacturer’s instructions. The purity of the isolated RNA was evaluated with optical density ratios (A260/A280 and A260/A230), using the NanoPhotometer (Implen, Munich, Germany). The RNA integrity was assessed with RNA integrity numbers (RIN) and DV200 using the RNA Nano 6000 Assay Kit and Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Library synthesis was performed on samples with DV200 greater than 20%. The RNA sequencing library was constructed using the TruSeq RNA Access Library Kit (Illumina, San Diego, CA, USA), followed by sequencing using the HiSeq2500 sequencing system (Illumina).
Analysis of RNA sequencing data
We used FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to confirm the read quality. Alignment to the human genome GRCh37 was performed using STAR-2.5.2a (https://github.com/alexdobin/STAR/releases/tag/2.5.2a), and quality assessment of the binary-sequence alignment map (BAM) for each sample was performed using the Genomon 2 RNA analysis pipeline (https://github.com/Genomon-Project) at the Human Genome Center, the Institute of Medical Science, University of Tokyo (Tokyo, Japan). The mapped read count for each sample was calculated using HTseq (https://htseq.readthedocs.io/en/release_0.9.1/) and normalised. The obtained read count data of genes were subjected to principal component analysis (PCA), and identification of the differentially expressed genes (DEGs) between the groups was performed using “DESeq2” Bioconductor R package (v1.26.0; http://www.bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html).
The ingenuity pathway analysis (IPA; Ingenuity Systems, Redwood City, CA, USA) was used to identify and understand the biological significance of the pathways involving the DEG products. The IPA interrogated the identified DEGs as “focus genes” against genes curated in their ingenuity knowledge database based on functional annotations and experimental observations.
The correlations of mRNA expression for genes were estimated by Pearson correlation analysis. The RNA sequencing read count data for each gene, normalised using the “Relative Log Expression (RLE)” method implemented in the “DESeq2” R package (v1.26.0), were used to calculate the correlation coefficient (R) between genes using the statistical programming environment R 4.0.3 (R Project for Statistical Computing, Vienna, Austria. https://www.R-project.org/).
External datasets for RNA sequencing of lung adenocarcinomas
We obtained fastq files of RNA sequencing for 463 lung adenocarcinomas from The Cancer Genome Atlas (TCGA) under an authorized account. The data were analysed using the Genomon 2 RNA analysis pipeline as our original RNA sequencing data described above.
Immunohistochemical analysis
Immunohistochemical staining was performed for the formalin-fixed paraffin-embedded tissue sections using an automated immunostainer (Histostainer 48A; Nichirei, Tokyo, Japan) with the following primary antibodies at respective dilutions: anti-KMT2C (Millipore Cat# ABE1851, dilution 1:300), anti-ATM (Abcam Cat# ab32420, RRID: AB_725574, dilution 1:100), anti-ATR (LSBio Cat# LS B4288-50, RRID: AB_10945735, dilution 1:50), anti-BRCA1 (Abcam Cat# ab16780, RRID: AB_2259338, dilution 1:200), anti-BRCA2 (Sigma-Aldrich Cat# HPA026815, RRID: AB_10602692, dilution 1:50), and anti-RAD51 (Abcam Cat# ab16780, RRID: AB_2722613, dilution 1:200). EnVison Flex+ Mouse LINKER (Agilent Technologies, Santa Clara, CA, USA) was added to amplify the signal of the anti-ATR antibody. The expression of each marker was detected using the Histofine Simple Stain MAX-PO (M) or (R) kits (Nichirei, Tokyo, Japan). Appropriate positive and negative control tissues were used in each experiment. Each staining was classified by a score from 0 to 3+ based on the proportion and intensity of the immunoreactive tumour cells (score 0: absent or weak nuclear expression in <50% of tumour cells, score 1+: weak nuclear expression in ≥50% of tumour cells, score 2+: moderate nuclear expression in ≥50% of tumour cells, and score 3+: strong nuclear expression in ≥50% of tumour cells).
Statistical analyses
The threshold value to identify the DEGs was a greater than 2-fold change in expression, with a P value <0.05 and an adjusted P value <0.05. In IPA analysis, we evaluated the probability of the identified pathways by calculating the P-value using the right-tailed Fisher’s exact test, and pathways with a P value <0.05 were considered significant. Additionally, z-scores were automatically calculated by the IPA, and positive and negative z-scores indicated activation and inactivation of pathways, respectively. Z-scores for pathways involving DEGs without enough information regarding their biological significance in the IPA dataset were ineligible for assignment and were designated as NaN.
The Mann-Whitney U test was performed to compare the scores of immunohistochemical analysis. Spearman’s correlation analysis was performed to examine the correlation between the immunohistochemical scores of KMT2C and each HRR factor. The correlation strength with respect to the correlation coefficient was determined as follows: very weak correlation at |r|<0.2, weak correlation at 0.2≤|r|<0.4, moderate correlation at 0.4≤|r|<0.6, strong correlation at 0.6≤|r|<0.8, and very strong correlation at 0.8≤|r|. In all statistical tests, P values <0.05 were considered significant. Statistical analyses were performed using EZR (Easy R) statistical software, version 1.36 (Saitama Medical Center, Jichi Medical University, Saitama, Japan) and a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Clinicopathological features
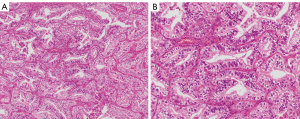
The clinicopathological findings for the lung cancer cases with and without an H-FLAC component are summarised in Table 1. In the cases with a H-FLAC component, columnar clear cells formed glandular, cribriform, and/or papillary structures with pseudostratified atypical nuclei, displaying a “piano keyboard-like” appearance (Figure 1). The H-FLAC components existed in various proportions: 5–9% in one case; 10–19% in two cases; 50–59% in one case; 60–69% in four cases; 70–79% in three cases; 80–89% in one case; 90–99% in two cases; and 100% in two cases. The predominant histology of the common lung adenocarcinoma cases was as follows: lepidic in two cases, acinar in five cases, papillary in seven cases, and solid in two cases.
Table 1
| Features | Lung cancers with H-FLAC | Common adenocarcinomas |
|---|---|---|
| Total number (n) | 16 | 16 |
| Age (year) | ||
| Median | 67 | 68 |
| Range | 52–81 | 55–78 |
| Sex | ||
| Female | 3 | 3 |
| Male | 13 | 13 |
| Smoking | ||
| Never | 2 | 2 |
| Ever | 14 | 14 |
| Stage | ||
| IA | 3 | 3 |
| IB | 5 | 5 |
| IIA | 3 | 3 |
| IIB | 2 | 2 |
| IIIA | 3 | 3 |
H-FLAC, high-grade fetal adenocarcinoma.
Transcriptome analysis using PCA, DESeq2, and IPA
RNA samples from 15 H-FLAC and all 16 common adenocarcinoma counterparts passed the quality inspection and were subjected to RNA sequencing. All the sequencing data acquired for each sample had an average of more than 25 million reads, and the quality of sequencing was over Q30, which was confirmed using FastQC. Differences in gene expression profile were first analysed by PCA (Figure S1), where H-FLAC was clearly distinguished from common adenocarcinoma regardless of an H-FLAC component ratio similar to the total tumour volume.
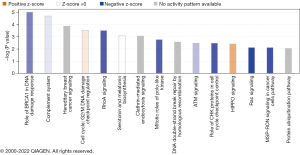
DESeq2 analysis identified 2112 genes that were significantly differentially expressed in H-FLAC and common adenocarcinoma. All DEGs identified are shown in available online: https://cdn.amegroups.cn/static/public/tlcr-23-137-1.xlsx. The DEGs were then used to identify the significant biological pathways associated with H-FLAC in comparison with common adenocarcinoma. Forty-six pathways were identified, 32 of which were inactivated in H-FLAC, 3 were activated, and the remaining 11 pathways were not assigned to either direction based on the z-score (Table S1). The pathway most significantly associated with H-FLAC was the “role of BRCA1 in DNA damage response”, which was inactivated in H-FLAC (Figure 2). Notably, multiple pathways related to DNA damage responses and cell cycle checkpoint regulations were significantly associated with H-FLAC. The pathways “ATM signalling” and “role of CHK proteins in cell cycle checkpoint control” were inactivated, whilst the pathways “G2/M DNA damage checkpoint regulation” and “p53 signalling” were activated (Figure 2, Figure S2). Although the direction of activation-inactivation was not assigned, the pathways “hereditary breast cancer signalling” and “DNA double-strand break repair by homologous recombination” showed significant association with H-FLAC, due to the DEGs involved in these pathways (Figure 2, Table S1). In addition to the DNA damage/cell cycle regulation-related pathways, multiple pathways related to small GTPases such as “RhoA signalling,” “Rac signalling,” and “signalling by Rho family GTPases” were inactivated in H-FLAC (Figure 2). Consistent with the inactivation of Rho/Rac signalling pathways, pathways “actin cytoskeleton signalling” and “paxillin signalling” were also inactivated in H-FLAC (Figure 2). Pathways with P values less than 0.01 [corresponding to −log (P value) greater than 2] are depicted in Figure 2. All the significant pathways are listed in Table S1 and are depicted in Figure S2.
We further analysed DEGs between six H-FLACs with KMT2C mutation and nine without mutations. Nineteen genes were identified as DEGs and their IPA analysis only found one pathway, nNOS (neuronal nitric oxides synthase) signalling in skeletal muscle cell without activity pattern (Figure S3A).
KMT2C expression and HRR genes in H-FLAC
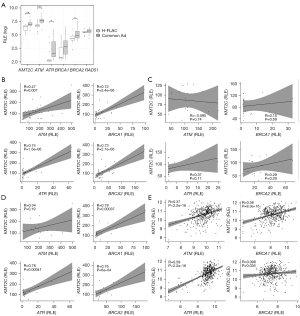
KMT2C expression in H-FLAC was significantly lower (less than a half) than that in common adenocarcinomas (log2 fold change =−1.09, P=1×10−4, available online: https://cdn.amegroups.cn/static/public/tlcr-23-137-1.xlsx). The expression of KMT2C, ATM, ATR, and BRCA2 was significantly lower in H-FLAC than in common adenocarcinomas (Figure 3A). Pearson correlation analysis for all cases revealed that KMT2C expression showed a strong positive correlation with ATR, BRCA1, and BRCA2 expression [correlation coefficient (R) greater than 0.7], and a moderately positive correlation with ATM expression (R=0.47; Figure 3B). Although H-FLAC did not show significant correlation between KMT2C and the expression of any of the HRR factors examined (Figure 3C), KMT2C expression showed a strong positive correlation with ATR, BRCA1, and BRCA2 expression in common adenocarcinomas (Figure 3D). Furthermore, the positive correlation between KMT2C and HRR genes ATM, ATR, and BRCA1 was observed in the 463 TCGA lung adenocarcinoma cases.
We further compared the expression of ATM, ATR, BRCA1, and BRCA2 in six H-FLACs with KMT2C mutation and nine without mutations. No genes showed significantly different expression between the two groups (Figure S3B).
Immunohistochemical analysis for KMT2C expression and HRR factors
The immunohistochemical features are displayed in Figures 4,5. In the F1, F3, F7, and F9 cases, non-fetal-type components could not be evaluated because the tumours were almost entirely comprised of H-FLAC components (Figure 4). Low KMT2C expression with a score of 1+ was identified in more H-FLAC components than in common adenocarcinomas (Figures 4,5A), and H-FLACs showed significantly lower KMT2C expression than in common adenocarcinomas (Table 2, Figure S4). Some cases showed lower KMT2C expression in the H-FLAC component than in the non-fetal-type component, although there were no significant differences between them (Table 2). Additionally, expression of HRR factors except RAD51 tended to be lower in H-FLAC components than in common adenocarcinomas (Figures 4,5B-5D), and ATM, ATR, and BRCA2 expression was significantly lower in H-FLAC components than in common adenocarcinomas (Table 2, Figure S4). Low nuclear expression (score 0 or 1+) of RAD51 was observed in 12/16 cases of H-FLACs and 12/16 cases of common adenocarcinomas (Figures 4,5F). Only one case (F7) showed cytoplasmic RAD51 expression. As the difference between the expression of fetal and non-fetal components of H-FLAC cases was unclear, only images of fetal components of the H-FLAC cases and cancer tissues of the common adenocarcinomas are shown in Figure 5.
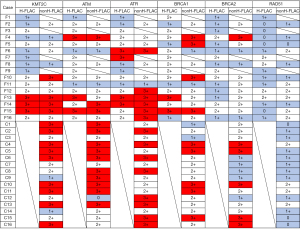
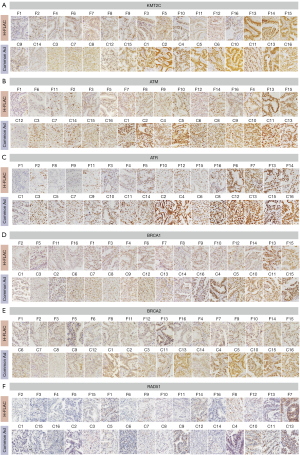
Table 2
| Factors | P value |
|---|---|
| KMT2C | |
| H-FLAC vs. nonH-FLAC | 0.44 |
| H-FLAC vs. commonAd | 0.02 |
| ATM | |
| H-FLAC vs. nonH-FLAC | >0.99 |
| H-FLAC vs. commonAd | 0.02 |
| ATR | |
| H-FLAC vs. nonH-FLAC | 0.17 |
| H-FLAC vs. commonAd | 0.03 |
| BRCA1 | |
| H-FLAC vs. nonH-FLAC | 0.27 |
| H-FLAC vs. commonAd | 0.18 |
| BRCA2 | |
| H-FLAC vs. nonH-FLAC | 0.04 |
| H-FLAC vs. commonAd | 0.03 |
| RAD51 | |
| H-FLAC vs. nonH-FLAC | 0.12 |
| H-FLAC vs. commonAd | 0.64 |
H-FLAC: the fetal-type component of the case F1–16; nonH-FLAC: the non-fetal-type adenocarcinoma component of the case F1–16; commonAd: common lung adenocarcinoma of the case C1–16. H-FLAC, high-grade fetal adenocarcinoma.
Next, we examined the correlation of the immunohistochemical scores between KMT2C and each HRR factor for all cases using Spearman’s correlation. The correlation coefficients between KMT2C and HRR factors were 0.553 (P=0.001) for ATM, 0.445 (P=0.01) for ATR, 0.378 (P=0.03) for BRCA1, 0.423 (P=0.02) for BRCA2, 0.117 (P=0.52) for RAD51. ATM, ATR, and BRCA2 expression moderately correlated with KMT2C expression, and BRCA1 expression weakly correlated with KMT2C expression. RAD51 expression showed no significant correlation with KMT2C expression. In the correlation analysis of H-FLACs or common adenocarcinomas, there was no significant correlation between the immunohistochemical scores of KMT2C and any of the HRR factors.
Discussion
Key findings
Our previous study identified missense or nonsense KMT2C mutations in six out of the 16 H-FLAC cases (38%) examined, whereas 14% of common lung adenocarcinomas harboured mutations in KMT2C according to the TumorPortal (http://www.tumorportal.org/) (11). Based on these findings, we surmised that KMT2C aberration may be a characteristic of H-FLAC. In this study, we revealed that both mRNA and protein expression of KMT2C were attenuated in H-FLAC. Further, HRR pathways were identified as significantly inactivated in H-FLACs by Ingenuity pathway analysis. Accordingly, the RNA and protein expression of ATM, ATR, and BRCA2 was significantly lower in H-FLAC components than in common adenocarcinomas. In addition, KMT2C expression was strongly correlated with that of ATR, BRCA1, and BRCA2 and was moderately correlated with that of ATM in adenocarcinomas together with H-FLAC and common adenocarcinoma or in common adenocarcinoma alone. The correlated expression of KMT2C and HRR genes examined seemed to be a characteristic of adenocarcinoma of the lung including ones with H-FLAC components.
Strengths and limitations
We analysed a cohort of rare lung cancer cases with H-FLAC components. To our knowledge, this is the first comprehensive expression analysis of H-FLAC cases. However, this study did have some limitations. The first issue is that we used a small control cohort of common lung adenocarcinoma cases for comparative analyses with H-FLAC. Although using a large-scale cohort is preferred, most publicly available expression datasets of lung adenocarcinomas (such as TCGA) are derived from frozen tissues instead of formalin-fixed paraffin-embedded tissues, and it was difficult to adopt such datasets for this study. Therefore, we used the control cohort of 16 cases (the same number of H-FLAC cases) that were matched based on clinicopathological factors. If more cases could have been analysed, a significant correlation between expressions of KMT2C and HRR factors might have been obtained after dividing the group into H-FLACs and common adenocarcinomas. However, due to the rarity of H-FLACs and the cost of analysis, it was difficult to increase the number of cases. The second is that we did not observe differences between H-FLAC with and without KMT2C mutations in expression of KMT2C and HRR genes examined and in the pathway analysis of differentially expressed genes. This result could be partly due to small number of cases analysed and may further raise a possibility that low expression of KMT2C was caused both genetically and epigenetically. The third limitation is that we did not directly clarify the KMT2C regulatory effect on the expression of the HRR factors in lung cancers with an H-FLAC component. Thus, further studies are required to determine this function of KMT2C in H-FLAC using assays such as chromatin immunoprecipitation. In the future, clinical studies will be required to assess the efficacy of PARP inhibitors in lung cancers, including H-FLAC, with KMT2C mutation/low expression.
Comparison with similar researches
KMT2C mutations have been identified in various malignant tumours. Although studies in mice showed a tumour suppressor role of KMT2C in acute myeloid leukaemia and urothelial tumorigenesis (21,22), little is known about the consequences of KMT2C dysfunction in cancer cells. Recently, Rampias et al. reported that KMT2C knockdown caused decreased expression of HRR factors such as ATM, ATR, BRCA1, and BRCA2 and increased sensitivity of urothelial cancer cells to the PARP inhibitor olaparib (13). In contrast, Chang et al. showed that KMT2C is recruited to the DNA damage site and directly mediates the HRR system through H3K4 methylation, chromatin relaxation, and secondary recruitment of HRR factors (23). It was also shown that both mutation and knockdown of KMT2C caused sensitization to olaparib in non-small cell lung cancer cells (23). These findings suggest that KMT2C directly and/or indirectly promotes HRR, and KMT2C dysfunction is associated with sensitivity to PARP inhibitors. In our present study, RNA sequencing and immunohistochemical analyses showed that H-FLAC components showed lower expression of KMT2C and HRR factors than common lung adenocarcinomas, and KMT2C expression was positively correlated with the expression of HRR factors. Notably, RNA sequencing analysis showed strong correlations between the expression of KMT2C and HRR factors exclusively in H-FLACs but not in common adenocarcinomas. Our findings support the role of KMT2C as a positive regulator for the expression of HRR factors, which is compromised in H-FLACs.
As opposed to ovarian and breast cancers, in which the efficacy of PARP inhibitors is well studied (24,25), very few studies explore their efficacy in non-small-cell lung cancers. It was recently reported that knockdown of ATM, BRCA1, and BRCA2 in non-small-cell lung cancer cells caused their hypersensitivity to olaparib (26). Although no clinical trials have demonstrated obvious therapeutic benefits of PARP inhibitors in non-small-cell lung cancers, a phase III trial of veliparib, a PARP inhibitor, in combination with platinum-based chemotherapy (NCT02106546) showed a favourable trend in lung squamous cell carcinomas with an LP52 signature, based on a 52-gene expression histology classifier (27). Hence, an appropriate case selection may contribute to the therapeutic benefits of PARP inhibitors in non-small-cell lung cancers. As H-FLACs rarely exhibit druggable mutations and PD-L1 expression (11), PARP inhibitors could be good therapeutic candidates for treatment of H-FLAC. Although there are few studies on RAD51 expression in lung cancers, high RAD51 expression was reported to be correlated with resistance to PARP inhibitors in germline BRCA-mutated breast cancers and ovarian cancers (28,29). The low RAD51 expression of H-FLACs may be a predictive finding for sensitivity to PARP inhibitors.
Although it was reported that inactivating mutations in HRR genes are associated with a high tumour mutation burden in breast cancer (30), H-FLACs showed a high tumour mutation burden in only 3/16 cases in our previous study (11). Therefore, unlike mutations, decreased expression of HRR genes may not contribute to an increase in mutation burden in H-FLAC. However, two of the three H-FLACs with a high tumour mutation burden exhibited low immunohistochemical expression (score 1+) of KMT2C, and the additional case harboured a nonsynonymous KMT2C mutation. Further studies of larger cohorts are required to elucidate an association between KMT2C mutation/low expression and tumour mutation burden in H-FLAC.
Our transcriptome analysis suggested that the Rho/Rac signalling pathway is inactivated as well. Rho/Rac GTPases regulate various cellular functions, including cell polarity, cell migration, and cell division (31). Notably, Rho GTPases play a key role in cancer cell invasion through dynamic regulation of actin cytoskeleton remodelling and cell adhesion. Recurrent gain-of-function mutations on RhoA have been reported in diffuse-type gastric carcinomas, characterised by a highly malignant phenotype with infiltrative invasion (32). However, the effect of Rho pathway inactivation on non-small-cell lung cancer has been unclear. Rho pathway inactivation might contribute to the formation of the fetal lung-like morphology and/or complex glandular structure in H-FLACs.
Explanations of findings
This study showed that KMT2C expression was significantly lower in H-FLACs than in common adenocarcinomas and was positively correlated with the expression of HRR factors.
Implications and actions needed
Our previous (11) and present studies suggest that the H-FLACs may be characterised by a high frequency of mutations and/or low expression of KMT2C. We speculate that H-FLACs with a high degree of KMT2C mutation/low expression may exhibit high sensitivity to PARP inhibitors. As we did not directly clarify the KMT2C regulatory effect on HRR factor expression, further studies are required to determine this function of KMT2C in H-FLAC using assays such as chromatin immunoprecipitation. Due to its rarity, more clinical studies of H-FLAC may not be realistic, but studies to assess the efficacy of PARP inhibitors in lung cancers based on stratification with KMT2C mutation/low expression are needed to improve therapeutic strategies and benefit H-FLAC patients.
Conclusions
H-FLAC components showed lower expression of KMT2C and HRR factors than common lung adenocarcinomas. KMT2C expression was positively correlated with the expression of HRR factors in lung adenocarcinomas. These results support the hypothesis that loss of KMT2C function decreases the expression of the HRR factors in H-FLACs. We speculated that PARP inhibitors could be an effective drug for treatment of H-FLACs with KMT2C mutation/low expression.
Acknowledgments
We are grateful to the technical staff at Kanagawa Cancer Center for their expert technical assistance and the members of the Clinical Sequencing Team at Human Genome Center, Institute of Medical Science, The University of Tokyo, for their expert technical assistance and intellectual input. The super-computing resource was provided by the Human Genome Center, the Institute of Medical Science, The University of Tokyo (http://sc.hgc.jp/shirokane.html). Genomon 2 (https://github.com/Genomon-Project) was provided by Kenichi Chiba, Ai Okada, and Yuichi Shiraishi.
Funding: This work was supported in part by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-137/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-137/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-137/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-137/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Kanagawa Cancer Center (approval No. H28-Res60) in Yokohama, Japan, and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nakatani Y, Kitamura H, Inayama Y, et al. Pulmonary adenocarcinomas of the fetal lung type: a clinicopathologic study indicating differences in histology, epidemiology, and natural history of low-grade and high-grade forms. Am J Surg Pathol 1998;22:399-411. [Crossref] [PubMed]
- WHO Classification of Tumours Editorial Board. Thoracic Tumours. 5th ed. Lyon: International Agency for Research on Cancer, 2021.
- Sato S, Koike T, Yamato Y, et al. Resected well-differentiated fetal pulmonary adenocarcinoma and summary of 25 cases reported in Japan. Jpn J Thorac Cardiovasc Surg 2006;54:539-42. [Crossref] [PubMed]
- Morita S, Yoshida A, Goto A, et al. High-grade lung adenocarcinoma with fetal lung-like morphology: clinicopathologic, immunohistochemical, and molecular analyses of 17 cases. Am J Surg Pathol 2013;37:924-32. [Crossref] [PubMed]
- Zhang J, Sun J, Liang XL, et al. Differences between low and high grade fetal adenocarcinoma of the lung: a clinicopathological and molecular study. J Thorac Dis 2017;9:2071-8. [Crossref] [PubMed]
- Suzuki M, Yazawa T, Ota S, et al. High-grade fetal adenocarcinoma of the lung is a tumour with a fetal phenotype that shows diverse differentiation, including high-grade neuroendocrine carcinoma: a clinicopathological, immunohistochemical and mutational study of 20 cases. Histopathology 2015;67:806-16. [Crossref] [PubMed]
- Suzuki M, Nakatani Y, Ito H, et al. Pulmonary adenocarcinoma with high-grade fetal adenocarcinoma component has a poor prognosis, comparable to that of micropapillary adenocarcinoma. Mod Pathol 2018;31:1404-17. [Crossref] [PubMed]
- Nakatani Y, Masudo K, Miyagi Y, et al. Aberrant nuclear localization and gene mutation of beta-catenin in low-grade adenocarcinoma of fetal lung type: up-regulation of the Wnt signaling pathway may be a common denominator for the development of tumors that form morules. Mod Pathol 2002;15:617-24. [Crossref] [PubMed]
- de Kock L, Bah I, Wu Y, et al. Germline and Somatic DICER1 Mutations in a Well-Differentiated Fetal Adenocarcinoma of the Lung. J Thorac Oncol 2016;11:e31-3. [Crossref] [PubMed]
- Li Y, Xi SY, Yong JJ, et al. Morphologic, Immunohistochemical, and Genetic Differences Between High-grade and Low-grade Fetal Adenocarcinomas of the Lung. Am J Surg Pathol 2021;45:1464-75. [Crossref] [PubMed]
- Suzuki M, Kasajima R, Yokose T, et al. Comprehensive molecular analysis of genomic profiles and PD-L1 expression in lung adenocarcinoma with a high-grade fetal adenocarcinoma component. Transl Lung Cancer Res 2021;10:1292-304. [Crossref] [PubMed]
- Hu D, Gao X, Morgan MA, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 2013;33:4745-54. [Crossref] [PubMed]
- Rampias T, Karagiannis D, Avgeris M, et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep 2019;20:e46821. [Crossref] [PubMed]
- Heale JT, Ball AR Jr, Schmiesing JA, et al. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell 2006;21:837-48. [Crossref] [PubMed]
- Rouleau M, Patel A, Hendzel MJ, et al. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010;10:293-301. [Crossref] [PubMed]
- Alexander M, Kim SY, Cheng H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020;198:897-907. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Wang Z, Jia R, Wang L, et al. The Emerging Roles of Rad51 in Cancer and Its Potential as a Therapeutic Target. Front Oncol 2022;12:935593. [Crossref] [PubMed]
- Brierley JD, Gospodarowicz MK, Wittekind Ch. TNM Classification of Malignant Tumors. 8th ed. Hoboken, NJ: Wiley-Blackwell, 2017.
- Sobin LH, Gospodarowicz MK, Wittekind Ch. TNM Classification of Malignant Tumors. 7th ed. Hoboken, NJ: Wiley-Blackwell, 2009.
- Chen C, Liu Y, Rappaport AR, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 2014;25:652-65. [Crossref] [PubMed]
- Lee J, Kim DH, Lee S, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci U S A 2009;106:8513-8. [Crossref] [PubMed]
- Chang A, Liu L, Ashby JM, et al. Recruitment of KMT2C/MLL3 to DNA Damage Sites Mediates DNA Damage Responses and Regulates PARP Inhibitor Sensitivity in Cancer. Cancer Res 2021;81:3358-73. [Crossref] [PubMed]
- Mirza MR, Coleman RL, González-Martín A, et al. The forefront of ovarian cancer therapy: update on PARP inhibitors. Ann Oncol 2020;31:1148-59. [Crossref] [PubMed]
- Zimmer AS, Gillard M, Lipkowitz S, et al. Update on PARP Inhibitors in Breast Cancer. Curr Treat Options Oncol 2018;19:21. [Crossref] [PubMed]
- Ji W, Weng X, Xu D, et al. Non-small cell lung cancer cells with deficiencies in homologous recombination genes are sensitive to PARP inhibitors. Biochem Biophys Res Commun 2020;522:121-6. [Crossref] [PubMed]
- Ramalingam SS, Novello S, Guclu SZ, et al. Veliparib in Combination With Platinum-Based Chemotherapy for First-Line Treatment of Advanced Squamous Cell Lung Cancer: A Randomized, Multicenter Phase III Study. J Clin Oncol 2021;39:3633-44. [Crossref] [PubMed]
- Cruz C, Castroviejo-Bermejo M, Gutiérrez-Enríquez S, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol 2018;29:1203-10. [Crossref] [PubMed]
- Feng Y, Wang D, Xiong L, et al. Predictive value of RAD51 on the survival and drug responsiveness of ovarian cancer. Cancer Cell Int 2021;21:249. [Crossref] [PubMed]
- Mei P, Freitag CE, Wei L, et al. High tumor mutation burden is associated with DNA damage repair gene mutation in breast carcinomas. Diagn Pathol 2020;15:50. [Crossref] [PubMed]
- Clayton NS, Ridley AJ. Targeting Rho GTPase Signaling Networks in Cancer. Front Cell Dev Biol 2020;8:222. [Crossref] [PubMed]
- Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583-7. [Crossref] [PubMed]


