
Machine learning model for circulating tumor DNA detection in chronic obstructive pulmonary disease patients with lung cancer
Highlight box
Key findings
• In chronic obstructive pulmonary disease patients with lung cancer, circulating tumor DNA (ctDNA) mutations were likely to be observed in those with advanced clinical stage, high C-reactive protein level, and milder emphysema. This finding was validated in machine learning models with high accuracy.
What is known and what is new?
• It is known that factors including tumor size, stage, metastasis, histology, and tumor necrosis are predictive of ctDNA shedding.
• An inverse relationship between emphysema and ctDNA detection is novel finding of this study. An increase of 1% in the emphysema index was associated with a 7% decrease in the odds of ctDNA detection.
What is the implication, and what should change now?
• Although patients with severe emphysema are in great need for non-invasive diagnosis of lung cancer, ctDNA detection might have limited clinical utility in this population.
Introduction
Lung cancer often develops in patients with an underlying pulmonary disease. Of them, chronic obstructive pulmonary disease (COPD) is the most common disease, along with pulmonary fibrosis (1,2). Studies have documented that COPD is an established risk factor for lung cancer development, even in never-smokers or when smoking exposure is controlled (3-9). When lung cancer is radiologically suspected, patients with COPD have a higher likelihood of being diagnosed with lung cancer and have higher rates of complications from invasive procedures than those without COPD (10). In this context, some COPD patients with lung cancer do not receive histologic diagnosis even when the tumor stage is I or II (11), necessitating a non-invasive biomarker that could aid lung cancer diagnosis in this high-risk population.
Cell-free DNA (cfDNA) is a non-encapsulating DNA in the peripheral blood, which was first discovered in 1948 (12). Many types of tumors release small DNA fragments through a combination of apoptosis, necrosis, and secretion (13). Although circulating tumor DNA (ctDNA) comprises only a small fraction of the total blood cfDNA (0.01% to 10%), the genomic alterations of ctDNA are highly specific to those of the original tumor and have been widely studied along with the development of next-generation sequencing (NGS). Analyses of ctDNA in plasma are intensively studied in patients with advanced-stage lung cancer to guide and monitor genotype-directed therapies (14,15). However, attempts to integrate ctDNA analysis into the diagnosis of lung cancer have been faced with challenges (16,17). One of the major limitations is that not all tumors shed sufficient amounts of ctDNA into the peripheral circulation in earlier stages (18). Given the relatively low sensitivity and high cost of ctDNA analysis, it is important to identify patients who are more likely to benefit from it. Several predictors of ctDNA shedding have been established, including tumor size, stage, number and sites of metastasis, non-adenocarcinoma histology, and tumor necrosis (17-20). However, previous studies included lung cancer patients regardless of underlying COPD, and little is known about the clinical factors associated with ctDNA detection in COPD patients with lung cancer.
Thus, we analyzed ctDNA mutations in spirometry-confirmed COPD patients with newly diagnosed lung cancer and sought to investigate which COPD-related clinical and imaging variables are associated with ctDNA detection. In addition, we developed prediction models to identify patients who are most likely to benefit from ctDNA analysis using multivariable machine learning (ML) methods. We present this article in accordance with the STARD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-633/rc).
Methods
A detailed description of the methods can be found in Appendix 1.
Study population
From October 2017 to September 2020, 461 patients with spirometry-defined COPD [post-bronchodilator forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC) <0.7] aged ≥40 years were prospectively enrolled from a single referral hospital. All of them did not have a significant pulmonary fibrosis. After excluding patients whose blood samples did not pass quality control or had technical issues in sample processing or library preparation (N=43), who withdrew consent or did not collect blood samples (N=11), and who had a history of malignancy other than lung cancer (N=3), 404 patients were included in the study population. For the present study, we further excluded patients without lung cancer (N=209), those with missing variables (N=10), and never smokers (N=8). Finally, 177 COPD patients with newly diagnosed lung cancer were included in the analysis. This study was approved by the Institutional Review Board (IRB) of Samsung Medical Center (IRB file No. SMC 2017-08-128). In addition, surgically resected lung cancer tissues from three patients were banked and provided by the Samsung Medical Center Biobank (IRB file No. SMC 2020-12-016). Informed consent was obtained from all the patients and the study was carried out in accordance with the Declaration of Helsinki (as revised in 2013).
Sequencing data processing and somatic mutation calling
Details of the sample preparation, DNA extraction and library preparation were described in the Appendix 1. For the library construction of plasma cfDNA, hybrid selection was performed using three customized baits (LungCancer v1, LiquidSCAN v2-PanCancer, or IVD v1.0, GENINUS, Seoul, Korea; Table S1). Each capture bait targeted 36, 38, and 46 cancer-related genes and covered 340, 117, and 174 kb genomic regions across the human genome.
All liquid biopsy sequencing data were aligned to the hg19 reference using BWA-mem (v0.7.5). GATK v4.0.0 (21) and SAMTOOLS v1.6 (22) were used for base quality recalibration and cross-validation of the unique molecular identifier (UMI) family, and for sorting SAM and BAM files, respectively. After sequencing alignment, discordant paired and off-target sequencing reads were removed. Picard (v2.9.4) was used to group reads into the same UMI families and in-house python (v2.7.10) scripts were used for error suppression. The error suppression method was based on previous studies (20) with minor modifications. First, all bases were subjected to Phred quality filtering using a threshold Q of 30 and only positions where total depths were above 500× were considered for variant identification. To exclude germline mutations in the analysis, non-reference alleles present at a frequency greater than 1% in the matched white blood cell gDNA were removed. The error suppression method using UMIs was used to distinguish true somatic mutations from polymerase chain reaction (PCR) and sequencing errors. After applying the error suppression method to the sequencing data, the following selection steps were used to eliminate the remaining sequencing errors: (I) variants not significantly greater than the error found in the matched germline DNA (binomial Bonferroni-adjusted P<0.01) were filtered out; (II) variant candidates with a high strand bias (90% if supporting reads ≥20; Fisher’s exact test, P<0.1 if supporting reads <20) were removed; (III) if the z-statistic of the variants was not significantly higher than the background error obtained from gDNA (Bonferroni-adjusted P<0.05), they were excluded from the analysis.
Finally, the mutation candidates were selected according to the following conditions: Allele frequencies ≥0.15% and alternative allele counts ≥5 were selected. For tissue specimens, somatic variants were identified using different criteria: total depth ≥100× and allele frequency ≥2%. In the case of insertions or deletions, variants with an allele frequency ≥5% were selected. Variants were annotated using variant effect predictor (VEP) (v102) (23) and nonsynonymous variants were used in this analysis.
Clinical variables
Demographic and clinical information were obtained from electronic medical records, including age, sex, body mass index (BMI), smoking status, tumor stage (24) and centrality (25). Regarding COPD, modified Medical Research Council (mMRC) grade, COPD assessment test (CAT), pulmonary function tests (26,27), and chest CT parameters were collected. Using automatic segmentation software (Aview, Coreline Soft, Seoul, Korea) (28,29), we measured the emphysema index (EI), defined as the percentage of lung area with CT attenuation values <−950 HU in the whole lung at inspiration. WBC count and high-sensitivity C-reactive protein (hsCRP) levels were also measured in blood samples.
Statistical analysis
Logistic regression (LR) analyses were performed to analyze the clinical factors associated with the detection of ctDNA. In multivariable LR models (Models 1–5), we used a panel as an adjusted variable. To estimate the prediction score of ctDNA detection in COPD patients, we used the sum of beta coefficients of significant variables from Model 5. To predict ctDNA detection using the variables, we considered four binary classifying machine learning (ML) models [logistic regression (LR), elastic net logistic regression (EN), random forest (RF), and support vector machine (SV)]. After splitting the dataset into training and test sets within the frame of leave-one-out cross-validation, we selected variables as features for ML models that showed significant association (P<0.1) with the presence of ctDNA mutation in a univariable LR model within each training set. The hyperparameters for EN, RF, and SV models were optimized by using grid search 5 cross-validation for accuracy in each training set. EN model was tuned by alpha from 0.0001 to 100, and L1 ratios between 0.0 and 1. RF model was allowed to have 10 to 1,000 estimators, maximum depth between 6 and 12, minimum samples per leaf between 8 and 18, and minimum samples per split between 8 and 20. SV model was allowed to use either radial or linear kernels, with gamma and C parameters between 0.001 to 100. To evaluate each model, we estimated the area under the receiver operating characteristics (ROC) curve (AUC), accuracy, sensitivity, specificity, and positive predictive value in the test set, and represented the performance of each model using an ROC curve plot. The model with the highest AUC was selected as the best prediction model for the ctDNA detection.
Results
Clinical characteristics of COPD patients with lung cancer
The clinical variables of patients with COPD and treatment-naïve lung cancer (N=177) are summarized in Table 1. Overall, the mean (standard deviation, SD) age was 69.8 (6.7) years, and most patients were male (94.4%) with former (67.8%) or current (32.2%) smoking exposure. Symptomatic burden measured using the mMRC dyspnea scale and CAT was relatively mild. Pulmonary function tests showed that the mean FEV1 was 70.3% pred, and the majority of patients (90.4%) had FEV1 ≥50% pred. The mean (SD) EI was 4.43% (6.8%) and 24 (13.6%) patients had an EI ≥10%. The clinical stages of lung cancer were classified as stage I (41.8%), II (17.5%), III (29.9%), and IV (10.7%), respectively. There were eight patients whose lung cancers were not histologically confirmed, mainly due to poor lung function, although the clinical diagnosis of lung cancer was unequivocal. Most patients (92.9%) had non-small cell lung cancer (NSCLC) and adenocarcinoma accounted for 40% of NSCLC cases.
Table 1
| Clinical variables | Overall (N=177) | ctDNA not detected (n=123) | ctDNA detected (n=54) | Univariable OR (95% CI) |
P |
|---|---|---|---|---|---|
| Age (years) | 69.8 (6.7) | 70.3 (6.6) | 68.5 (6.8) | 0.96 (0.91–1.01) | 0.082 |
| Sex, male | 167 (94.4) | 115 (93.5) | 52 (96.3) | 1.81 (0.43–12.26) | 0.440 |
| Smoking | |||||
| Former | 120 (67.8) | 88 (71.5) | 32 (59.3) | Reference | |
| Current | 57 (32.2) | 35 (28.5) | 22 (40.7) | 1.73 (0.88–3.38) | 0.111 |
| BMI (kg/m2) | 23.2 (2.8) | 23.3 (2.7) | 23.1 (3.0) | 0.98 (0.87–1.10) | 0.731 |
| mMRC ≥2 | 48 (27.1) | 26 (21.1) | 22 (40.7) | 2.56 (1.28–5.16) | 0.008 |
| CAT total ≥10 | 99 (55.9) | 65 (52.8) | 34 (63.0) | 1.52 (0.79–2.96) | 0.21 |
| Pulmonary function | |||||
| FVC, % pred | 88.9 (13.7) | 89.0 (14.6) | 88.7 (11.6) | 1.00 (0.98–1.02) | 0.907 |
| FEV1, % pred | 70.3 (15.4) | 69.7 (15.7) | 71.5 (14.7) | 1.01 (0.99–1.03) | 0.489 |
| FEV1/FVC, % | 55.6 (10.1) | 54.8 (10.3) | 57.5 (9.3) | 1.03 (1.00–1.07) | 0.089 |
| FEV1 <50% pred, n | 17 (9.6) | 13 (10.6) | 4 (7.4) | 0.68 (0.18–2.02) | 0.502 |
| EI | |||||
| % total lung | 4.43 (6.8) | 5.07 (7.2) | 2.98 (5.6) | 0.94 (0.88–1.00) | 0.041 |
| ≥10%, n | 24 (13.6) | 21 (17.1) | 3 (5.6) | 0.29 (0.07–0.88) | 0.027 |
| % tumor-located lobe (N=175) | 4.24 (8.2) | 5.11 (9.5) | 2.22 (3.4) | 0.92 (0.83–0.98) | 0.010 |
| CRP (mg/dL) | 1.06 (1.9) | 0.66 (1.4) | 2.00 (2.6) | 1.41 (1.18–1.72) | <0.001 |
| Tumor size (mm) | 37.2 (19.5) | 32.1 (15.1) | 48.9 (23.1) | 1.05 (1.03–1.07) | <0.001 |
| Clinical stage of lung cancer | |||||
| I | 74 (41.8) | 68 (55.3) | 6 (11.1) | Reference | |
| II | 31 (17.5) | 23 (18.7) | 8 (14.8) | 3.94 (1.24–13.15) | 0.020 |
| III | 53 (29.9) | 25 (20.3) | 28 (51.9) | 12.69 (4.98–37.32) | <0.001 |
| IV | 19 (10.7) | 7 (5.7) | 12 (22.2) | 19.43 (5.85–73.40) | <0.001 |
| Centrally located tumor | 71 (40.1) | 39 (31.7) | 32 (59.3) | 3.13 (1.63–6.14) | <0.001 |
| Histology (N=169) | |||||
| NSCLC | 157 (92.9) | 112 (97.4) | 45 (83.3) | Reference | |
| SCLC | 12 (7.1) | 3 (2.6) | 9 (16.7) | 7.47 (2.12–34.82) | <0.001 |
| Histology of NSCLC (N=157) | |||||
| Non-adenocarcinoma | 94 (59.9) | 66 (58.9) | 28 (62.2) | Reference | |
| Adenocarcinoma | 63 (40.1) | 46 (41.1) | 17 (37.8) | 0.87 (0.42–1.76) | 0.703 |
| Sequencing panel | |||||
| IVDv1 | 82 (46.3) | 58 (47.2) | 24 (44.4) | Reference | |
| LCv1 | 41 (23.2) | 27 (22.0) | 14 (25.9) | 1.25 (0.55–2.78) | 0.583 |
| PCv2 | 54 (30.5) | 38 (30.9) | 16 (29.6) | 1.02 (0.47–2.15) | 0.964 |
Values indicate the number of number (%) or mean (standard deviation) for categorical and continuous variables, respectively. COPD, chronic obstructive pulmonary disease; ctDNA, circulating tumor DNA; OR, odds ratio; CI, confidence interval; BMI, body mass index; mMRC, modified medical research council; CAT, COPD assessment test; FVC, forced vital capacity; FEV1, forced expiratory volume in 1 second; EI, emphysema index; CRP, c-reactive protein; NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer.
Detection of ctDNA mutations in overall study population
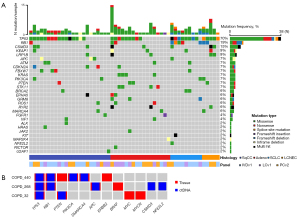
Among the 177 patients with COPD and treatment-naïve lung cancer, at least one ctDNA mutation was detected in 54 patients (30.5%). Detection rate was 8.1%, 25.8%, 52.8%, and 63.2% in stage I, II, III, and IV, respectively. The median number of detected mutations per patient was 2 (range, 1–8) and the median VAF of the mutations was 6.0% (range, 0.7–85.3%). The most frequently mutated genes were TP53 (70%), RB1 (19%), CSMD3 (15%), KEAP1 (9%), and LRP1B (9%) (Figure 1A). TP53 was the most frequently mutated gene in both adenocarcinoma (52.9%) and squamous cell carcinoma (69.6%). Among the 54 patients, 19 underwent surgical resection without neoadjuvant treatment. Tumor tissues and adjacent normal lung tissues were banked in three patients. To confirm that ctDNA mutations identified by our pipeline were derived from tumor tissues, we compared ctDNA mutations and mutations identified in tumor tissues of the same patient (Figure 1B). All ctDNA mutations (16 mutations within 6 genes) were also detected in tumor tissues across the three patients, while 62.5% (10/16) of tumor tissue mutations were detected in ctDNA, suggesting that ctDNA mutations are derived from the tumor tissues and can be used in the subsequent analyses as tumor mutations in COPD patients with lung cancer.
Clinical factors associated with ctDNA detection
To identify ctDNA detection-associated factors, we first compared the variables of patients with ctDNA detection (N=54) and those without ctDNA detection (N=123) using univariable models (Table 1). As a result, patients with ctDNA mutations (N=54) had higher mMRC grade, lower EI, higher CRP, larger tumor size, more advanced clinical stages, more centrally located tumors, and a higher prevalence of small cell lung cancer (SCLC) than patients without ctDNA detection (Table 1). As different sequencing panels were used in our mutation data, we further conducted multivariable LR analyses with the same variables adjusted for sequencing panel type (Table 2), considering different types of EI (continuous variable in Model 2, binary variable using cut-off of 10% in Model 3, or continuous variable of tumor located lobes in Model 4). Tumor stages were most strongly associated with ctDNA detection in all the models {adjusted odds ratio (OR) comparing stage II, III, and IV to stage I: 3.82 [95% confidence interval (CI): 1.14–13.58], 9.01 (95% CI: 3.23–28.61), and 15.52 (95% CI: 4.15–66.14), respectively, in Model 2}. Lower EI values of the total lung and higher CRP levels were also significantly associated with ctDNA detection. In Model 2, a 1% increase in EI of the total lung was associated with a 7% decrease in the odds of ctDNA detection (adjusted OR: 0.933, 95% CI: 0.857–0.999, P=0.047).
Table 2
| Clinical variables | Model 1* | Model 2† | Model 3† | Model 4† | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||||
| Age | 0.96 (0.91–1.01) | 0.081 | |||||||||
| Sex | 1.73 (0.41–11.8) | 0.482 | |||||||||
| Smoking | |||||||||||
| Former | Reference | ||||||||||
| Current | 1.71 (0.86–3.38) | 0.126 | |||||||||
| BMI (kg/m2) | 0.98 (0.87–1.10) | 0.710 | |||||||||
| mMRC ≥2 | 2.57 (1.27–5.23) | 0.009 | 2.05 (0.88–4.79) | 0.095 | 1.97 (0.86–4.52) | 0.108 | 2.09 (0.89–4.93) | 0.091 | |||
| CAT total ≥10 | 1.50 (0.78–2.94) | 0.221 | |||||||||
| FEV1 <50% pred | 0.67 (0.18–2.00) | 0.484 | |||||||||
| EI | |||||||||||
| % total lung | 0.94 (0.88–1.00) | 0.043 | 0.93 (0.86–0.999) | 0.047 | |||||||
| ≥10%, n | 0.29 (0.07–0.89) | 0.029 | 0.29 (0.06–1.07) | 0.064 | |||||||
| % tumor located lobe | 0.92 (0.83–0.98) | 0.009 | 0.93 (0.83–1.00) | 0.061 | |||||||
| CRP (mg/dL) | 1.41 (1.18–1.74) | <0.001 | 1.39 (1.12–1.78) | 0.002 | 1.42 (1.14–1.83) | 0.001 | 1.39 (1.11–1.79) | 0.003 | |||
| Clinical stage of lung cancer | |||||||||||
| I | Reference | Reference | Reference | Reference | |||||||
| II | 3.96 (1.25–13.23) | 0.020 | 3.82 (1.14–13.58) | 0.029 | 4.10 (1.24–14.41) | 0.021 | 3.55 (1.06–12.59) | 0.040 | |||
| III | 12.87 (4.99–38.30) | <0.001 | 9.01 (3.23–28.61) | <0.001 | 10.17 (3.75–31.67) | <0.001 | 8.10 (2.90–25.71) | <0.001 | |||
| IV | 19.16 (5.75–72.59) | <0.001 | 15.52 (4.15–66.14) | <0.001 | 17.43 (4.73–73.20) | <0.001 | 14.23 (3.63–63.06) | <0.001 | |||
| Centrally located tumor | 3.14 (1.63–6.17) | 0.001 | 1.75 (0.79–3.86) | 0.164 | 1.77 (0.80–3.93) | 0.160 | |||||
| Sequencing panel | |||||||||||
| IVDv1.0 | – | Reference | Reference | Reference | |||||||
| LCv1 | 0.78 (0.28–2.09) | 0.620 | 0.77 (0.28–2.06) | 0.602 | 0.83 (0.30–2.25) | 0.719 | |||||
| PCv2 | 0.88 (0.34–2.25) | 0.790 | 0.92 (0.36–2.32) | 0.864 | 0.87 (0.33–2.22) | 0.767 | |||||
*, adjusted only for sequencing panels. †, variables with P<0.05 from Model 1 were used in forward selection. The selected variables were then used in the construction of Models 2, 3, and 4. For the EI, continuous and binary values of EI of the total lung areas were used in Models 2 and 3, respectively. Continuous values of EI of the tumor located in lobes were used in Model 4. In Model 4, 175 samples were used as described in Table 1. ctDNA, circulating tumor DNA; COPD, chronic obstructive pulmonary disease; OR, odds ratio; CI, confidence interval; BMI, body mass index; mMRC, modified medical research council; CAT, COPD assessment test; FEV1, forced expiratory volume in 1 second; EI, emphysema index; CRP, C-reactive protein.
Prediction of ctDNA detection using machine learning models
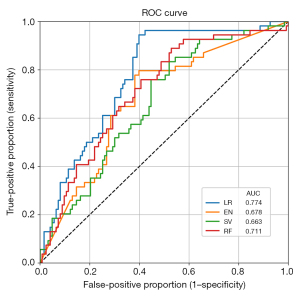
To predict ctDNA mutation detection using multiple variables, we selected variables with P<0.1 from the univariable LR models as the features of four ML prediction models (LR, EN, RV, SV) (Table 3 and Table S2). As shown in Figure 2, the LR model showed the highest AUC (0.774) with an accuracy of 71.8%, sensitivity of 42.6%, and specificity of 84.6% for predicting the presence of ctDNA mutations. We further estimated the prediction score per sample to show the effect of the significant variables on the risk of ctDNA detection in COPD patients with lung cancer using the beta coefficients of the multivariable LR model (Model 5, composed of variables with P<0.05 of the Model 2 adjusted by panel). After classifying samples based on the risk scores, we found that 82.4% of the patients in the highest (10th) decile group had ctDNA mutations while all patients in the lowest (1st) decile group had no ctDNA mutations (Figure 3 and Table S3).
Table 3
| Performance | LR | EN | SV | RF |
|---|---|---|---|---|
| Accuracy (%) | 71.8 | 65.5 | 71.8 | 70.1 |
| Specificity (%) | 84.6 | 72.4 | 94.3 | 92.7 |
| Sensitivity (%) | 42.6 | 50.0 | 20.4 | 18.5 |
| PPV (%) | 54.8 | 44.3 | 61.1 | 52.6 |
| AUC | 0.774 | 0.678 | 0.663 | 0.711 |
AUC, area under the receiver-operating-characteristics curve; ctDNA, circulating tumor DNA; EN, elastic net regression; LR, logistic regression; PPV, positive predictive value; RF, random forest; SV, support vector machine.

Prognostic values of ctDNA detection
During the median follow-up of 20.7 (interquartile range, 10.9–31.8) months, 51 (28.8%) patients with lung cancer died. The proportion of patients who died was significantly higher in those with ctDNA detection than in those without ctDNA detection (51.9% vs. 18.7%, P<0.001). In an unadjusted Cox regression model, ctDNA detection was associated with an increased risk of death [unadjusted hazard ratio (HR): 3.27, 95% CI: 1.87–5.72; Figure S1). However, after adjustment for major confounders, including tumor stage and histology, this association was not statistically significant. In a subgroup of patients with stage I and II lung cancer (N=105), ctDNA detection was independently associated with increased mortality (fully adjusted HR: 7.91, 95% CI: 1.55–40.36). VAF (per 1% increase) was also significantly associated with an increased risk of death in patients with stage I and II lung cancer (fully adjusted HR: 1.25, 95% CI: 1.01–1.56) (Table S4).
Discussion
Despite having a high risk of developing lung cancer, patients with COPD experience higher rates of complications from invasive diagnostic procedures compared to those without COPD. To focus on these high-risk populations, this study exclusively included patients with COPD with newly diagnosed lung cancer and analyzed ctDNA mutations using targeted deep sequencing. At least one ctDNA mutation was detected in 30.5% of patients (8.1% in stage I to 63.2% in stage IV). Of the comprehensively collected clinical and imaging variables, advanced clinical stage, lesser degree of emphysema, and increased CRP levels were associated with ctDNA detection among COPD patients with lung cancer. While this finding must be further validated, ML models with cross-validation demonstrated a satisfactory performance in identifying patients with ctDNA mutations, suggesting a potential clinical utility of ctDNA analysis assisted by a prediction model. We also confirmed that ctDNA detection and VAF levels were prognostic factors for poor overall survival (OS), particularly in early stage lung cancer patients.
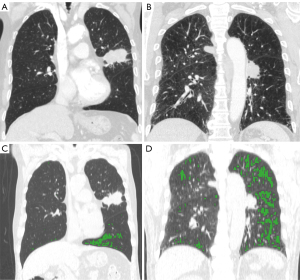
We found an inverse relationship between emphysema and ctDNA detection, which is a novel finding. Figure 4 shows representative cases of two patients with similar smoking exposure and lung function and the same stage IIB squamous cell carcinomas, which were located centrally. However, one patient with an EI of 1% had ctDNA mutations detected (RB1 and TP53) whereas the other patient, with an EI of 10%, was negative for ctDNA mutation. Given that the major process of ctDNA shedding is tumor cell apoptosis and release into the bloodstream, it might be attributable to impaired pulmonary vasculature in the emphysematous lung. Earlier histological studies reported vascular alterations in emphysema (30). Another study showed that endothelial dysfunction (decreased expression of VEGF) is associated with the extent of emphysema (31). Indeed, the cross-sectional area of small pulmonary vessels is inversely correlated with the extent of emphysema (32). Thus, the lower rate of ctDNA shedding in patients with severe emphysema in this study might be due to loss of vessels. This inverse correlation between emphysema and ctDNA detection in COPD patients with lung cancer suggests that ctDNA might have limited clinical utility in patients with severe emphysema who are in greater need for non-invasive diagnosis of lung cancer. Negative ctDNA mutation in this population cannot exclude lung cancer diagnosis. Therefore, it should be used as complementary to other modalities, such as chest CT scans.
Despite the limitations regarding insufficient shedding of ctDNA, several factors related to ctDNA detection have been reported. This study showed that advanced clinical stage is strongly associated with ctDNA detection in COPD patients with lung cancer, which was consistent with the correlation between tumor stage and ctDNA detection in many previous studies, as well as the number of metastatic sites (19). Based on several studies using NGS of multiple recurrent genetic alterations in lung cancer for the purpose of non-invasive diagnosis or residual disease detection (16,20,33,34), the sensitivity for stages II and III was up to 100% but the sensitivity was 50% or less for stage I NSCLC (16,34). Tumor size has been consistently associated with ctDNA shedding and a minimum tumor volume of 10 cm3, which corresponds to a nodule diameter of 2.6 cm (T1c stage), is required to quantify VAF of 0.1% (20). In addition, 18F-FDG avidity or metabolic tumor volume on positron emission tomography-CT scans were positively associated with ctDNA detection and VAF levels (17,20,35). Other radiologic parameters associated with the ctDNA detection rate include necrosis and nodule density (17). Among histological parameters, non-adenocarcinoma histology, SCLC, Ki67 proliferation index, necrosis, and lymphovascular invasion are known to predict ctDNA detection (20,36,37). This study did not include histological parameters in the multivariable models because we aimed to determine clinical factors predicting ctDNA detection before or even without histological confirmation as patients with COPD often have a high complication risk of invasive procedures.
Regarding the prognostic value of ctDNA detection, this study confirmed the findings of previous studies by showing that ctDNA detection and VAF levels are associated with shorter OS, particularly in early stages (17,38-40). Poor survival with positive ctDNA in early-stage lung cancer might stem from the higher recurrence rate after the surgery. Numerous studies have shown that preoperative and postoperative ctDNA detection was associated with shorter recurrence-free survival and OS after curative surgery (41-43). Accordingly, previous studies suggested the presence of ctDNA mutations from liquid biopsy into cancer staging as TNM “B” tumor staging, as ctDNA detection may reflect the presence of micrometastasis or minimal residual disease beyond a mere reproduction of information from tissues (44). Moreover, recent studies have shown that specific mutational profile or tumor mutational burden in ctDNA can also predict poor clinical outcome with polyclonal metastasis pattern and treatment response to immune checkpoint inhibitors (45,46).
In addition, the most frequently mutated genes in our data were also significantly mutated in lung adenocarcinoma (TP53, RB1, and KEAP1) and lung squamous cell carcinoma (TP53 and RB1) in a previous large-scale study using whole exome sequencing (47). Similarly, TP53 was the most frequently mutated gene in both lung adenocarcinoma (52.9% vs. 54.1%; our study vs. Campbell et al. (47) and lung squamous cell carcinoma (69.6% vs. 86.4%).
The strength of this study is the use of data from targeted deep sequencing and the adoption of ML models to predict ctDNA detection in an individual patient with COPD and lung cancer. The model can assist in non-invasive lung cancer diagnosis by estimating a probability of ctDNA detection. For example, based on the prediction scores, more than 82% of patients with the top 10% score had ctDNA mutations, suggesting that the diagnosis of lung cancer can be established using ctDNA in these patients. On the other hand, for some patients with low prediction scores (low probability of ctDNA detection), clinicians should also utilize other diagnostic tests rather than solely rely on the ctDNA analysis. In addition, this study only included patients with spirometry-confirmed COPD, who are at a higher risk of developing lung cancer compared to matched smokers, and collected comprehensive information regarding COPD, such as COPD symptoms, lung function, and quantitatively measured emphysema on CT.
This study also has several limitations. First, it was conducted in a single referral center and the study results were not externally validated. To address this limitation, we adopted machine learning models with cross-validation. In addition, as this study focused on COPD patients recruited from pulmonology clinics, our cohort predominantly consisted of men and smokers (>90%), which is consistent with the multicenter studies from Korea that are based on pulmonology clinics (48,49). This may limit the generalizability of our findings to other populations. As all patients were current or former smokers, the relatively lower prevalence of EGFR mutations in our cohort might be attributable to smoking and COPD (36,50). The lack of never-smokers made it impossible to explore the association between smoking exposure and ctDNA detection. Similarly, due to the unavailability of occupational information, the association between occupational exposure and ctDNA detection was not investigated. Second, genotyping in this study was limited to the pre-determined genes that were included in the panel, which are relatively fewer in number compared to previous studies (17,33). Thus, ctDNA detection might be underestimated compared to targeted sequencing with more genes, whole exome, or whole genome sequencing. Moreover, due to the difference in the genes between the panels used, the data regarding individual mutational features were not included in the current analysis. Nevertheless, we used panels as an adjusted covariate in multivariable models to minimize the effect of different panels on the outcomes. Finally, the mutations between ctDNA and tumor tissues were compared in only three patients. However, considering that all mutations from ctDNA were detected in the tumor tissues, which is consistent with previous reports (17,51,52), it was appropriate to use our ctDNA mutation data as a surrogate for tumor mutation data from the other patients in our analyses.
Conclusions
Using NGS of targeted genes, this study showed that approximately one-third of COPD patients shed ctDNA at the time of lung cancer diagnosis. In addition to the well-known correlation with the tumor stages, we found that patients with severe emphysema were less likely to have ctDNA detected, despite the presence of lung cancer. We also constructed ML models to predict ctDNA detection with high accuracy. Further studies incorporating individual mutational features and detailed radiologic parameters are needed to improve the prediction model for ctDNA detection and to develop prediction models for lung cancer diagnosis in COPD patients.
Acknowledgments
Funding: This research was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-633/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-633/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-633/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-633/coif). M.J.A. served as an unpaid editorial board member of Translational Lung Cancer Research from October 2021 to September 2023. Seung-Ho Shin and D.P. were employees, and W.Y.P. is the CEO and stakeholder of Geninus Inc. D.P. is also an employee of Planit Healthcare Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the IRB of Samsung Medical Center (IRB file no. SMC 2017-08-128). In addition, surgically resected lung cancer tissues from three patients were banked and provided by the Samsung Medical Center Biobank under approval of IRB of Samsung Medical Center (IRB file no. SMC 2020-12-016). Informed consent was obtained from all the patients including the consent for publication and the study was carried out in accordance with the Declaration of Helsinki (as revised in 2013). The representative case images were considered anonymized as identifying information was not included.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moro-Sibilot D, Aubert A, Diab S, et al. Comorbidities and Charlson score in resected stage I nonsmall cell lung cancer. Eur Respir J 2005;26:480-6. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Non-Small Cell Lung Cancer, version 7.2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (cited Nov 26, 2021).
- Tockman MS, Anthonisen NR, Wright EC, et al. Airways obstruction and the risk for lung cancer. Ann Intern Med 1987;106:512-8. [Crossref] [PubMed]
- de Torres JP, Bastarrika G, Wisnivesky JP, et al. Assessing the relationship between lung cancer risk and emphysema detected on low-dose CT of the chest. Chest 2007;132:1932-8. [Crossref] [PubMed]
- Turner MC, Chen Y, Krewski D, et al. Chronic obstructive pulmonary disease is associated with lung cancer mortality in a prospective study of never smokers. Am J Respir Crit Care Med 2007;176:285-90. [Crossref] [PubMed]
- Wilson DO, Weissfeld JL, Balkan A, et al. Association of radiographic emphysema and airflow obstruction with lung cancer. Am J Respir Crit Care Med 2008;178:738-44. [Crossref] [PubMed]
- Young RP, Hopkins RJ, Christmas T, et al. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur Respir J 2009;34:380-6. [Crossref] [PubMed]
- Park HY, Kang D, Shin SH, et al. Chronic obstructive pulmonary disease and lung cancer incidence in never smokers: a cohort study. Thorax 2020;75:506-9. [Crossref] [PubMed]
- Loganathan RS, Stover DE, Shi W, et al. Prevalence of COPD in women compared to men around the time of diagnosis of primary lung cancer. Chest 2006;129:1305-12. [Crossref] [PubMed]
- Iaccarino JM, Silvestri GA, Wiener RS. Patient-Level Trajectories and Outcomes After Low-Dose CT Screening in the National Lung Screening Trial. Chest 2019;156:965-71. [Crossref] [PubMed]
- Kang N, Shin SH, Noh JM, et al. Treatment modality and outcomes among early-stage non-small cell lung cancer patients with COPD: a cohort study. J Thorac Dis 2020;12:4651-60. [Crossref] [PubMed]
- MANDEL P. METAIS P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil 1948;142:241-3.
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020;580:245-51. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577-86. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987-93. [Crossref] [PubMed]
- McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Shin SH, Jeong DY, Lee KS, et al. Which definition of a central tumour is more predictive of occult mediastinal metastasis in nonsmall cell lung cancer patients with radiological N0 disease? Eur Respir J 2019;53:1801508. [Crossref] [PubMed]
- Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J 2005;26:319-38. [Crossref] [PubMed]
- Choi JK, Paek D, Lee JO. Normal predictive values of spirometry in Korean population. Tuberc Respir Dis 2005;58:230-42.
- Cho YH, Seo JB, Lee SM, et al. Quantitative CT Imaging in Chronic Obstructive Pulmonary Disease: Review of Current Status and Future Challenges. J Korean Soc Radiol 2018;78:1-12.
- Koo HJ, Lee SM, Seo JB, et al. Prediction of Pulmonary Function in Patients with Chronic Obstructive Pulmonary Disease: Correlation with Quantitative CT Parameters. Korean J Radiol 2019;20:683-92. [Crossref] [PubMed]
- Wright JL, Lawson L, Paré PD, et al. The structure and function of the pulmonary vasculature in mild chronic obstructive pulmonary disease. The effect of oxygen and exercise. Am Rev Respir Dis 1983;128:702-7. [Crossref] [PubMed]
- Kasahara Y, Tuder RM, Cool CD, et al. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am J Respir Crit Care Med 2001;163:737-44. [Crossref] [PubMed]
- Matsuoka S, Washko GR, Dransfield MT, et al. Quantitative CT measurement of cross-sectional area of small pulmonary vessel in COPD: correlations with emphysema and airflow limitation. Acad Radiol 2010;17:93-9. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Hyun MH, Lee ES, Eo JS, et al. Clinical implications of circulating cell-free DNA quantification and metabolic tumor burden in advanced non-small cell lung cancer. Lung Cancer 2019;134:158-66. [Crossref] [PubMed]
- Zhang Y, Yao Y, Xu Y, et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat Commun 2021;12:11. [Crossref] [PubMed]
- Cho MS, Park CH, Lee S, et al. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS One 2020;15:e0230622. [Crossref] [PubMed]
- Cargnin S, Canonico PL, Genazzani AA, et al. Quantitative Analysis of Circulating Cell-Free DNA for Correlation with Lung Cancer Survival: A Systematic Review and Meta-Analysis. J Thorac Oncol 2017;12:43-53. [Crossref] [PubMed]
- Isaksson S, George AM, Jönsson M, et al. Pre-operative plasma cell-free circulating tumor DNA and serum protein tumor markers as predictors of lung adenocarcinoma recurrence. Acta Oncol 2019;58:1079-86. [Crossref] [PubMed]
- Jee J, Lebow ES, Yeh R, et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat Med 2022;28:2353-63. [Crossref] [PubMed]
- Li N, Wang BX, Li J, et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer 2022;128:708-18. [Crossref] [PubMed]
- Xia L, Mei J, Kang R, et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin Cancer Res 2022;28:3308-17. [Crossref] [PubMed]
- Chen D, Guo J, Huang H, et al. Prognostic value of circulating tumor DNA in operable non-small cell lung cancer: a systematic review and reconstructed individual patient-data based meta-analysis. BMC Med 2023;21:467. [Crossref] [PubMed]
- Yang M, Forbes ME, Bitting RL, et al. Incorporating blood-based liquid biopsy information into cancer staging: time for a TNMB system? Ann Oncol 2018;29:311-23. [Crossref] [PubMed]
- Wang Z, Duan J, Cai S, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol 2019;5:696-702. [Crossref] [PubMed]
- Abbosh C, Frankell AM, Harrison T, et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 2023;616:553-62. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Lee JY, Chon GR, Rhee CK, et al. Characteristics of Patients with Chronic Obstructive Pulmonary Disease at the First Visit to a Pulmonary Medical Center in Korea: The KOrea COpd Subgroup Study Team Cohort. J Korean Med Sci 2016;31:553-60. [Crossref] [PubMed]
- Park TS, Lee JS, Seo JB, et al. Study Design and Outcomes of Korean Obstructive Lung Disease (KOLD) Cohort Study. Tuberc Respir Dis (Seoul) 2014;76:169-74. [Crossref] [PubMed]
- Chen H, Wang A, Wang J, et al. Target-based genomic profiling of ctDNA from Chinese non-small cell lung cancer patients: a result of real-world data. J Cancer Res Clin Oncol 2020;146:1867-76. [Crossref] [PubMed]
- Heeke S, Hofman V, Ilié M, et al. Prospective evaluation of NGS-based liquid biopsy in untreated late stage non-squamous lung carcinoma in a single institution. J Transl Med 2020;18:87. [Crossref] [PubMed]
- Schwaederlé MC, Patel SP, Husain H, et al. Utility of Genomic Assessment of Blood-Derived Circulating Tumor DNA (ctDNA) in Patients with Advanced Lung Adenocarcinoma. Clin Cancer Res 2017;23:5101-11. [Crossref] [PubMed]


