
Expression of cytokines in pleural effusions and corresponding cell lines of small cell lung cancer
Highlight box
Key findings
• The establishment of pleural small cell lung cancer (SCLC) cell lines allowed for the determination of the fraction of cytokines secreted by the tumor cells compared to the total cytokines found in malignant pleural effusions (MPEs).
What is known and what is new?
• The expression of vascular endothelial growth factor A (VEGFA), angiopoietins, osteopontin and other mediators have been reported previously for MPEs but this study reports the overexpression of protumor and immunosuppressive cytokines secreted by the pleural SCLC cells.
What is the implication, and what should change now?
• The analysis of cytokines expressed by the SCLC cells in MPEs provides a repertoire of cell-enriched mediators as potential targets for therapeutic intervention apart from VEGFA.
Introduction
Approximately 10–15% of patients with lung cancer are affected by malignant pleural effusions (MPEs) at the initial diagnosis and the median survival of these patients is as short as 3–12 months (1-3). The incidence of MPE may rise to 50% for cases at advanced stages (4). Pleural invasion was first reported in 1958 by Brewer et al. to be a negative predictor of survival for lung cancer (5). In detail, the median survival was 11.2, 5.9, and 4.8 months in patients with no pleural effusion, minimal pleural effusion and MPE, respectively. Due to the inferior overall survival (OS), MPE was reclassified to M1a (stage IV) category in the TNM system (6,7). Among malignancies, lung cancer is the most common cause of MPE and a 16% MPE rate was found in 57,685 patients with non-small cell lung cancer (NSCLC) at first presentation (8,9). It is well-known that “wet” pleural carcinomatosis has a poorer prognosis in comparison to “dry” pleural carcinomatosis (10). For SCLC, MPE was present in 7,639 (11.16%) of 68,443 patients with an estimated 1-year survival 17% vs. 30% without MPE (8,11). In another investigation, 1,770 primary SCLC patients showed pleural involvement (PI) in 25.4% of cases again constituting a negative prognostic factor (2). The analysis of 358 SCLC patients with extensive-stage disease and MPE revealed that 43.8% of patients died within 12 months (12). However, for most SCLC studies, the effect of PI on survival has not been considered (13).
Symptoms in most patients with MPE comprise dyspnea, cough and chest pain. In normal physiological conditions, there is 0.26 mL/kg of fluid in the pleural space and influx and exit of pleural space fluids are balanced to keep the volume constant (14). Excess fluid formation in the pleural space can be either due to malignant or benign causes (15). Fluid leaves the pleural space through the parietal pleura stomata, that are openings between mesothelial cells and fluid is drained by collecting lymphatics to the mediastinal lymph nodes (16). MPE occurs mostly because of impaired lymphatic drainage along this outward flow. Meyer et al. found a significant relation between mediastinal lymph node impairment and development of MPE (17). Tumor cells infiltrate the pleural space via the hematogenous, direct or lymphatic invasion and the lymphatic drainage can be blocked by growing tumor cells (18). However, only 55–60% of patients with pleural or lymphatic metastases develop MPE (19).
A variety of cells is present in the pleural microenvironment, including mesothelial and endothelial cells as well as cells of myeloid origin and of the lymphatic system. In general, the immune system effector cells interact with tumor cells to increase angiogenesis, inflammation and vascular leakiness eventually leading to the development of MPE (15,19). The immunoregulatory mediators present include IL-2, tumor necrosis factor (TNF) and interferons. Vascular permeability is promoted by vascular endothelial growth factor A (VEGFA) and matrix metalloproteinases (MMPs) by disruption of endothelial cell integrity and cellular junctions (20-22). Likewise, angiopoietin 1/2, secreted by tumor cells, participates in increased vascular permeability (23).
For the present investigation, MPEs of three advanced SCLC patients were collected and a total of 105 cytokines analyzed by a Western blot array using the Proteome Profiler Human XL Cytokine Array Kits. Furthermore, corresponding SCLC cancer lines were established in tissue culture of the same MPEs and subjected to the same cytokine analysis to determine the fraction of cytokines secreted by the cancer cells themselves in the MPE. In addition, the effect of the MPE samples on the proliferation of four permanent SCLC cell lines were measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. We present this article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-569/rc).
Methods
Pleural effusions and blood samples
Thoracocentesis of pleural effusions has been routinely performed for SCLC patients and the samples were obtained according to the guidelines set forth in the Ethics Approval 366/2003 granted by the Ethics Committee of the Medical University of Vienna, Vienna, Austria, including informed consent of the patients. The same protocol was valid for the acquisition of blood samples and the establishment of cell lines. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
Pleural effusions were centrifuged and the supernatants aliquoted and stored frozen at −80 ℃. The cells were washed with tissue culture medium (RPMI-1640 medium supplemented with 10% fetal bovine serum and antibiotics). All chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA) except indicated otherwise. Cells were kept under tissue culture conditions (5% CO2, 37 ℃) for short-term cultures at least until two 75 cm2 tissue culture flasks exhibited a complete coverage of the surface area before the conditioned media was used for the analysis of the cytokines. NCI-H69 cells were obtained from the ATCC (Manassa, VA, USA) and the SCLC lines SCLC26A, S457 and the SCLC CTC cell line BHGc40 have been established at our institution. The SCLC26A cell line was obtained from a patient prior to treatment, the S457 cell line from a patient progressing after second-line therapy with cyclophosphamide/epirubicin/vincristine (CEV) and BHGc40 from a blood sample of a patient also progressing after second-line CEV therapy.
Cytokine western blot arrays
Cytokines of pleural effusion samples or cell line supernatants were determined using Human Profiler Arrays Cytokine XL (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. The Western blot array spots were quantitated using ImageJ and Origin software. Tests were performed in duplicate and the six reference spots spotted to each membrane were used to calibrate the individual chemoluminescence intensities. The cytokines exhibiting highest expression were analyzed for overexpressed pathways using the Reactome Pathway database (reactome.org). Finally, the target effectors of these significantly altered pathways were summarized.
MTT tests
Cells (1×104) were distributed to the wells of 96 microtiter plates (TPP, Trasadingen, Switzerland) in 100 µL medium and supplemented 1:1 with twofold dilutions of pleural effusion samples. Viability of the cells was determined after 4 days incubation in tissue culture using a modified MTT test kit (EZ4U, Biomedica, Vienna, Austria). Tests were performed in triplicate with 10 dilutions, starting at 1:1 tissue culture medium: MPE, and three independent repetitions. The resulting data were calculated with Origin software (OriginLab, Northampton, MA, USA).
Statistical analysis
The Reactome analysis employs over-representation analysis (ORA) for checking in a dataset, whether the proportion of differentially expressed genes of a specific pathway exceeds the proportion of genes that could be randomly expected, by calculating the likelihood that the association between the sample identifiers and the found pathway is due to random chance. The statistical significance is calculated using the Binomial Test with P values (P<0.05) and the false discovery rate (FDR) estimated by the Benjamini-Hochberg approach, to identify false positive results.
Results
Cytokine profile of the three MPEs
The Proteome Profiler Human XL Cytokine Array Kit detects 105 human cytokines simultaneously using Western blot technology. Pleural effusion samples of three advanced SCLC patients (S996, S1033, S1035) were collected prior to the initiation of chemotherapy and processed for determination of the cytokines by these arrays. Results were calibrated using six included reference spots. The cytokines exhibiting the highest expression for these three pleural effusions are shown in Figure 1 and the results revealed a highly similar quantitative expression for these three independent SCLC MPE samples. Data for all cytokines measured are presented in Table S1. A large range of mediators are expressed in MPEs, including angiogenic effectors, cell adhesion molecules, proliferation stimulators, proteases, interleukins, and urokinase plasminogen activator surface receptor (uPAR/CD87) and mediators of inflammation and chemoattractants.

Overexpressed pathways of the cytokines detected in SCLC MPE
The functional significance of the large number of cytokines detected was searched for their inclusion in overexpressed cellular pathways using the Reactome Pathway Database. All highly expressed cytokines as shown in Figure 1 were submitted to Reactome analysis. Pathways determined for most significant overexpression include IL-4, IL-13, IL-10, IL-18 and IL-33 signaling, as well as neutrophil degranulation, extracellular matrix (ECM) organization, cell adhesion molecules, activators of ATF4, IGFs, PK3/AKT signaling and unfolded protein response (UPR) among others (Table 1).
Table 1
| Reactome pathway | Entity/total | Ratio | P value |
|---|---|---|---|
| IL-4 and IL-13 signaling | 17/211 | 0.014 | 1.11e−16 |
| IL-10 signaling | 10/86 | 0.006 | 1.11e−11 |
| Neutrophil degranulation | 12/478 | 0.031 | 2.08e−06 |
| ATF4 activation ER | 4/34 | 0.002 | 2.14e−05 |
| Regulation IGF and IGFBPs | 6/127 | 0.008 | 3.03e−05 |
| PERK regulates gene expression | 4/42 | 0.003 | 4.85e−05 |
| Integrin cell surface interactions | 5/86 | 0.006 | 5.48e−05 |
| IL-33 signaling | 2/42 | 0.63e−04 | 1.69e−04 |
| Unfolded protein response (UPR) | 5/156 | 0.01 | 8.37e−04 |
| IL-18 signaling | 2/11 | 0.001 | 7.22e−04 |
| Extracellular matrix organization | 7/328 | 0.022 | 8.56e−04 |
| Senescence SASP | 4/90 | 0.006 | 8.64e−04 |
| RUNX3 regulation | 2/10 | 0.001 | 6.57e−04 |
| PI3K/AKT signaling | 4/129 | 0.008 | 0.003 |
| TP53 regulation | 2/18 | 0.001 | 0.003 |
Cytokines with high expression were subjected to pathway analysis and the most overexpressed pathway listed, according to the components/entities of specific cascades found (ratio of number of entities/total number of pathway members) and the P values. The FDR ranged from 1.8e−14 to 0.042 from pathways with highest to lowest P values. FDR, false discover rate; ER, endoplasmic reticulum; IGFBPs, insulin-like growth factor binding proteins; SASP, senescence-associated secretory phenotype; MPE, malignant pleural effusion.
The chain of mediators constituting these pathways end in effector molecules and mechanisms shown in Table 2. The Reactome analysis depicts the cascade of proteins comprising the respective pathways ending in specific terminal effectors.
Table 2
| Reactome pathway targets |
| IFNγ signaling |
| Platelet degranulation |
| IGF1 regulation |
| MET STAT3/5 |
| Flt3 signaling |
| TP53 repair |
| PI3K AKT activation |
| Insulin receptor signaling |
| IL-4 and IL-13 signaling |
| PTEN loss |
| ErbB4 activation |
| UPR/senescence |
The ultimate effectors of the pathways found overexpressed in Reactome analysis are summarized. UPR, unfolded protein response; MPE, malignant pleural effusion; SCLC, small cell lung cancer.
The cytokines with the highest expression are shown and the results revealed a similar quantitative expression for these three independent cell lines (Table 2). Data for all cytokines measured are presented in Table S1. Results were calibrated using six included reference spots. A large range of cytokines are expressed by the tumor cells, including angiogenic effectors, cell adhesion molecules, proliferative effectors, proteases, interleukins, uPAR and mediators of inflammation and chemoattractants.
Overexpressed pathways of the cytokines in cell line supernatants
The functional significance of these cellular cytokines has been searched for their inclusion in overexpressed cellular pathways using the Reactome Pathway Database. All highly expressed cytokines as shown in Figure 2 were submitted to Reactome analysis. Pathways determined for most significant overexpression include IL-4, IL-13, IL-10 and IL-33 signaling, as well as platelet degranulation, growth factors, cell adhesion molecules, activation of TPAF2, PK3/AKT signaling and senescence-associated secretory phenotype (SASP) among others (Table 3).

Table 3
| Reactome pathway | Entity | Ratio | P value |
|---|---|---|---|
| IL-10 signaling | 22/86 | 0.006 | 1.11e−16 |
| IL-4/IL-13 signaling | 14/211 | 0.014 | 4.27e−12 |
| TFAP2 (AP-2) regulation | 4/21 | 0.001 | 4.75e−06 |
| IGF and IGFBPs | 6/127 | 0.008 | 5.16e−05 |
| Integrin interactions | 5/86 | 0.006 | 8.60e−05 |
| Platelet degranulation | 6/141 | 0.009 | 9.14e−05 |
| IL-33 signaling | 2/4 | 2.63e−04 | 2.04e−04 |
| PI3K/AKT signaling | 5/129 | 0.008 | 5.51e−04 |
| Senescence SASP | 4/90 | 0.006 | 0.001 |
Cytokines with high expression were subjected to pathway analysis and the most overexpressed pathway listed, according to the components/entities of specific cascades found (ratio of number of entities/total number of pathway members) and the P values. The FDR ranged from 1.0e10−16 to 0.003 from pathways with highest to lowest P values. SASP, senescence-associated secretory phenotype; FDR, false discovery rate.
The overexpressed pathways detected in supernatants of the cell lines by Reactome analysis follow a chain of effectors that terminate in specific proteins (Table 4).
Table 4
| Pathway targets |
| Neutrophil degranulation |
| Platelet activation |
| IGF1 |
| TP53 |
| IL-6, IL-4, IL-10, IL-13 |
| AP-2 regulation |
| Integrin cell surface |
| EGFR PI3K AKT |
| Pou5F1 SOX2 Nanog repress differentiation |
| VENTX regulation |
| UPR PERK ATF4 |
| PDGF |
The ultimate effectors of the pathways found in Reactome analysis for the three established SCLC cell lines are summarized. UPR, unfolded protein response; SCLC, small cell lung cancer.
Ratios of cytokine expression in cell line supernatants/expression in MPEs
The cytokines determined for the MPEs and cell lines were compared and the ratios of the expression calculated (Table 5). In particular, cellular DKK-1, ENA-78/CXCL5, G-CSF, GROa/CXCL1, MMP9, PF-4/CXCL4, RANTES/CCL5, TARC/CCL17 and VEGFA exhibited much higher concentrations compared to the levels found in the MPEs.
Table 5
| Gene | Ratio |
|---|---|
| BDNF | 1.93 |
| DKK-1 | 8.03 |
| ENA-78 CXCL5 | 18.80 |
| G-CSF | 5.28 |
| GM-CSF | 2.43 |
| GROa CXCL1 | 6.10 |
| IL-16 | 1.68 |
| IL-33 | 3.07 |
| IP-10 CXCL10 | 3.71 |
| I-TAC CXCL11 | 7.27 |
| LIF | 8.30 |
| MCP-1 | 1.63 |
| MCP-3 CCL7 | 9.77 |
| M-CSF | 1.98 |
| MIG CXCL9 | 4.00 |
| MIP 3a | 8.54 |
| MMP-9 | 3.40 |
| PF4 CXCL4 | 13.42 |
| RANTES CCL5 | 5.10 |
| TARC CCL17 | 6.46 |
| Thrombospondin-1 | 2.30 |
| TNF-a | 3.32 |
| uPAR | 2.38 |
| VEGFA | 31.23 |
MPE, malignant pleural effusion.
Effects of the MPE samples on proliferative activity of SCLC cell lines
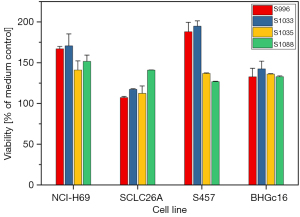
In order to investigate the potential effects of the MPE samples on the growth of a number of permanent SCLC cell lines, namely NCI-H69, SCLC26A, S457 and BHGc16, cells were supplemented with samples of four MPEs in proliferation tests (Figure 3). All MPE samples were found to stimulate the proliferation of all 4 cell lines in a dose-dependent manner. Figure 3 shows the results obtained with a 25% MPE fraction in tissue culture medium. The lowest medium response was determined for the SCLC26A cell line, derived from an SCLC patient without pretreatment. The SCLC S457 and the BHGc16 SCLC CTC lines have been established from progressing and chemoresistant SCLC patients employing a MPE and a blood sample, respectively.
Discussion
MPEs appear in all cell types of lung cancer with highest incidence in adenocarcinoma (approximately 30%), followed by squamous cell carcinoma (SCC; approximately 9%) and SCLC (approximately 10%) (24,25). Thoracocentesis of pleural effusion allows for the examination through cytology and proof of a malignant process (26). Accumulation of MPE is due to increased permeability of the pleural vessel cells as well as vasoactive and inflammatory cytokines secreted by invaded tumor cells. Even patients with minimal MPEs had a significantly increased risk of death with a hazard ratio of 1.454 (27). However, MPEs affect survival only in T2 stage but not in T3 and T4 stages. A worse prognosis associated with MPE seems to be linked to the high rate of lymph node metastasis in these patients and mediastinal lymphadenopathy is the main reason for the accumulation of pleural fluid (27,28). MPE in SCLC forms due to indirect infiltration of the lymphatic vessels (27). In general, the focus of the management MPE is palliative and without survival benefit.
The pleural mesothelial cell is a common cell in the pleural space and initiates responses to stimuli such as tumor cells (4,29). Malignant cells invade the parietal pleura and proliferate to form pleural metastases that secrete mediators such as VEGFA, chemokines, IL-6, osteopontin (OPN) and TNF provoking the accumulation of inflammatory cells (30,31). VEGFA stimulates proliferation and migration of endothelial cells and proved necessary for the accumulation of pleural fluid through increase of vascular permeability (32,33). Additionally, these cytokines have been reported to trigger in turn proinflammatory and proangiogenic mediators controlled by NF-κB and STAT3 (31,34). OPN seems to promote MPE by enhancing VEGFA release from vascular endothelial or mesothelial cells (35). Increases of the expression of ICAM-1 and VCAM-1 promote release of metalloproteinases by the cancer cells (36,37). In summary, different cell populations and molecules are involved in the formation of MPE either stimulating pleural inflammation (e.g., IL-2, TNF and IFN) or tumor angiogenesis [e.g., angiopoietin 1 (ANG-1), angiopoietin 2 (ANG-2)] and vascular hyperpermeability (e.g., VEGFA, MMP, MCP-1/CCL2, OPN, and others) (18,21). Our analysis of the cytokines expressed in MPEs and corresponding SCLC cell lines document presence of the cytokines cited above and a wide range of other mediators. The net effect of this host of cytokines seems to be the high stimulation of tumor growth in a suitable modified pleural environment.
The establishment of corresponding SCLC cell cultures from the same MPEs used for the measurement of cytokines allows to identify the fraction of mediators provided by the tumor cell populations for the first time. A large range of cytokines of the SCLC cell lines exhibit a relatively higher production compared to the matching MPE. Of these, high expression of MIP-3α/CCL20 recruits inflammatory cells and promotes lung cancer cell migration and proliferation through the activation of ERK and PI3K signaling pathways (38). The receptor for advanced glycation end (RAGE) products is known to induce the accumulation of tumor-associated macrophages (TAMs) in lung cancer tissue and to further accelerate the tumor progression (39). The higher concentrations of the epithelial neutrophil-activating peptide 78 (ENA-78) in MPEs can stimulate a progressive influx of neutrophils into the pleural space (40). Furthermore, CXCL9, also known as monokine-induced by interferon-γ (MIG), is secreted during inflammatory conditions and may act directly on tumor cells via their CXCR3 receptor to promote cell migration and epithelial mesenchymal transition (EMT) (41).
TARC/CCL17 is a ligand of CCR4 that induces chemotaxis of Th2 and inhibitory regulatory T (Treg) cells and, additionally, recruit eosinophils into the tumor (42). Tumor-derived GROa/CXCL1 contributes to tumor-associated neutrophil (TAN) infiltration in lung cancer which accelerates tumor growth (43). A correlation exists between the IL-16 concentrations and the number of CD4+ T-cells, and furthermore, IL-16 produces a significant influx of CD4+ T-cells into the pleural space (44). TNF binds to its receptor TNFR2 of tumor cells and enhances the suppressive activity of Tregs by up-regulating CTLA-4 and PD-L1 expression (45). Besides regulating angiogenesis and tumor vasculature, thrombospondin-1 limits antitumor immunity by CD47-mediated inhibition of innate and adaptive immune cells (46). Additionally, IP-10/CXCL10 is highly expressed by cancer cells and correlates with infiltration by Tregs and poor survival (47). Moreover, IP-10 signaling via its cognate receptor CXCR3 was shown to increase tumor growth, migration and invasion of cancer cells in various tumor types (48). The high concentrations of PF4 in MPE is linked with a poor prognosis, most likely by the suppression of T lymphocyte response that promotes tumor progression (49). The G-CSF- or GM-CSF-secreting cancers exhibit rapid progression due to a cytokine-mediated immune suppression and angiogenesis (50). These range of cytokines derived from cancer cells seem to promote tumor growth and development of MPE by the generation of a markedly immunosuppressive milieu in the pleural effusion (51).
The other cytokines overexpressed in the SCLC lines are involved in promotion of increased cellular signaling, tumor progression and invasion. High expression of VEGFA in conjunction with MMP-9 has been reported in pleural effusions of patients due to malignant diseases (52-54). In MPEs, VEGFA is the most important angiogenic factor and increases vascular permeability as well as migration of endothelial cells together with MMPs and other factors (20,21,52). Drugs targeting VEGFA are effective and safe for the clinical management of MPE (21). Among the interleukins, the alarmin IL-33 induces several factors that activate NF-κB, JNK, ERK, and p38, eventually leading to the expression of cytokines, chemokines, and growth factors (55). Overexpression of leukemia inhibitory factor (LIF) is linked to the characteristics of aggressive tumors including lymph node metastasis and progression and increases STAT3 phosphorylation (56). Brain-derived neurotrophic factor (BDNF) can increase cancer cell growth, survival, migration and anoikis, via TrkB and the p75NTR death receptor (57). The BDNF/TrkB pathways trigger downstream signaling, including PI3K/Akt, Jak/STAT, ERK, NF-kB, and activation of EGFR. CC chemokine ligand 7 (CCL7) and its receptors CCR1, CCR2 and CCR3 were found highly expressed during lung cancer bone metastasis (58). The expression of uPAR is increased during inflammation, especially in invasive tumors (59). uPAR expression plays a key role in tumorigenicity, tumor proliferation, invasion and glycolytic tumor metabolism. Monocyte chemoattractant protein-1 (MCP-1)/CCL2 is secreted from tumor cells and tumor stroma. The blockade of MCP-1, by neutralizing antibodies, has been demonstrated to suppress tumorigenesis in solid tumors (60). Dysregulated expression of CXCL5/ENA-78 has been shown to be involved in tumor metastasis and angiogenesis (61). Neutralizing CXCL5/ENA-78 with antibodies impairs cancer progression and increases the inhibitory effects of tyrosine kinase inhibitors (62). Furthermore, CXCL5 inhibition showed low side effects in experimental animals. In regard to PF4, antiplatelet factors with great potential for clinical application are studied but it proved difficult to reduce the side effects of such drugs (63). In conclusion, SCLC cells in MPEs produce cytokines that promote tumor growth and protect the malignant cells by impairing antitumor immune system responses.
Conclusions
We report for the first time a comparison of the cytokines expressed in MPEs with the same mediators released into supernatants of the corresponding SCLC cell lines. Thus, tumor-promoting factors and immunosuppressive mediators of the tumor cells could be separated from effectors supplied by nonmalignant cell populations of the pleural space.
Acknowledgments
We thank Dr. T. Hohenheim (retired) for continuing endorsement.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-569/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-569/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-569/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-569/coif). G.H. serves as an unpaid editorial board member of Translational Lung Cancer Research from September 2023 to August 2025. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Samples were obtained following the regulations of the Ethics Approval 366/2003 granted by the Ethics Committee of the Medical University of Vienna, Vienna, Austria. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Yang J, Yin H, Liu M, et al. Effect of pleural invasion on survival of patients with small cell lung cancer: Propensity score analysis and nomogram establishment based on the SEER database. Front Surg 2023;10:1108732. [Crossref] [PubMed]
- Bibby AC, Dorn P, Psallidas I, et al. ERS/EACTS statement on the management of malignant pleural effusions. Eur Respir J 2018;52:1800349. [Crossref] [PubMed]
- Feller-Kopman DJ, Reddy CB, DeCamp MM, et al. Management of Malignant Pleural Effusions. An Official ATS/STS/STR Clinical Practice Guideline. Am J Respir Crit Care Med 2018;198:839-49. [Crossref] [PubMed]
- Brewer LA, Bai AF, Little JN, et al. Carcinoma of the lung; practical classification for early diagnosis and surgical treatment. J Am Med Assoc 1958;166:1149-55. [Crossref] [PubMed]
- Morgensztern D, Waqar S, Subramanian J, et al. Prognostic impact of malignant pleural effusion at presentation in patients with metastatic non-small-cell lung cancer. J Thorac Oncol 2012;7:1485-9. [Crossref] [PubMed]
- Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest 1997;111:1710-7. [Crossref] [PubMed]
- Shojaee S, Singh I, Solsky I, et al. Malignant Pleural Effusion at Presentation in Patients with Small-Cell Lung Cancer. Respiration 2019;98:198-202. [Crossref] [PubMed]
- Skok K, Hladnik G, Grm A, et al. Malignant Pleural Effusion and Its Current Management: A Review. Medicina (Kaunas) 2019;55:490. [Crossref] [PubMed]
- Agalioti T, Giannou AD, Krontira AC, et al. Mutant KRAS promotes malignant pleural effusion formation. Nat Commun 2017;8:15205. [Crossref] [PubMed]
- Keidan N, Aujayeb A. Small Cell Lung Cancer and Pleural Effusion: An Analysis from a District General Hospital. Pulm Ther 2023;9:359-65. [Crossref] [PubMed]
- Penz E, Watt KN, Hergott CA, et al. Management of malignant pleural effusion: challenges and solutions. Cancer Manag Res 2017;9:229-41. [Crossref] [PubMed]
- Wang S, Yang L, Ci B, et al. Development and Validation of a Nomogram Prognostic Model for SCLC Patients. J Thorac Oncol 2018;13:1338-48. [Crossref] [PubMed]
- Feller-Kopman D, Light R. Pleural Disease. N Engl J Med 2018;378:740-51. [Crossref] [PubMed]
- Beaudoin S, Gonzalez AV. Evaluation of the patient with pleural effusion. CMAJ 2018;190:E291-5. [Crossref] [PubMed]
- Sahn SA. The pathophysiology of pleural effusions. Annu Rev Med 1990;41:7-13. [Crossref] [PubMed]
- Meyer PC. Metastatic carcinoma of the pleura. Thorax 1966;21:437-43. [Crossref] [PubMed]
- Psallidas I, Kalomenidis I, Porcel JM, et al. Malignant pleural effusion: from bench to bedside. Eur Respir Rev 2016;25:189-98. [Crossref] [PubMed]
- Stathopoulos GT, Kalomenidis I. Malignant pleural effusion: tumor-host interactions unleashed. Am J Respir Crit Care Med 2012;186:487-92. [Crossref] [PubMed]
- Hamed EA, El-Noweihi AM, Mohamed AZ, et al. Vasoactive mediators (VEGF and TNF-alpha) in patients with malignant and tuberculous pleural effusions. Respirology 2004;9:81-6. [Crossref] [PubMed]
- Chen Y, Mathy NW, Lu H. The role of VEGF in the diagnosis and treatment of malignant pleural effusion in patients with non-small cell lung cancer Mol Med Rep 2018;17:8019-30. (Review). [Crossref] [PubMed]
- Damianovich M, Hout Siloni G, Barshack I, et al. Structural basis for hyperpermeability of tumor vessels in advanced lung adenocarcinoma complicated by pleural effusion. Clin Lung Cancer 2013;14:688-98. [Crossref] [PubMed]
- Fang SC, Zhang HT, Hu HD, et al. Effect of Endostar combined with angiopoietin-2 inhibitor on malignant pleural effusion in mice. Med Oncol 2015;32:410. [Crossref] [PubMed]
- Medenica M, Medenica M, Cosovic D. Pleural Effusions in Lung Cancer: Detection and Treatment. In: Torres AFC. editor. Lung Cancer - Strategies for Diagnosis and Treatment. London: IntechOpen, 2018. DOI:
10.5772/intechopen.78307 . - Hsu LH, Hsu PC, Liao TL, et al. Pleural fluid osteopontin, vascular endothelial growth factor, and urokinase-type plasminogen activator levels as predictors of pleurodesis outcome and prognosticators in patients with malignant pleural effusion: a prospective cohort study. BMC Cancer 2016;16:463. [Crossref] [PubMed]
- Shahini L, Hoxha M, Marku F, et al. Role of cytoblock on pleural effusion for diagnosis of malignant disease. Diagn Cytopathol 2023;51:684-8. [Crossref] [PubMed]
- Ryu JS, Lim JH, Lee JM, et al. Minimal Pleural Effusion in Small Cell Lung Cancer: Proportion, Mechanisms, and Prognostic Effect. Radiology 2016;278:593-600. [Crossref] [PubMed]
- Wang F, Li P, Li F. Nomogram for Predicting the Relationship between the Extent of Visceral Pleural Invasion and Survival in Non-Small-Cell Lung Cancer. Can Respir J 2021;2021:8816860. [Crossref] [PubMed]
- Tsavlis D, Katopodi T, Anestakis D, et al. Molecular and Immune Phenotypic Modifications during Metastatic Dissemination in Lung Carcinogenesis. Cancers (Basel) 2022;14:3626. [Crossref] [PubMed]
- Sahn SA. Pleural diseases related to metastatic malignancies. Eur Respir J 1997;10:1907-13. [Crossref] [PubMed]
- Stathopoulos GT, Zhu Z, Everhart MB, et al. Nuclear factor-κB affects tumor progression in a mouse model of malignant pleural effusion. Am J Respir Cell Mol Biol 2006;34:142-50. [Crossref] [PubMed]
- Yano S, Herbst SH, Shinohara H, et al. Treatment for malignant pleural effusion of human lung adenocarcinoma by inhibition of vascular endothelial growth factor receptor tyrosine kinaze phosphorylation. Clin Cancer Res 2000;6:957-65.
- Thickett DR, Armstrong L, Millar AB. Vascular endothelial growth factor (VEGF) in inflammatory and malignant pleural effusions. Thorax 1999;54:707-10. [Crossref] [PubMed]
- Giannou AD, Marazioti A, Spella M, et al. Mast cells mediate malignant pleural effusion formation. J Clin Invest 2015;125:2317-34. [Crossref] [PubMed]
- Cui R, Takahashi F, Ohashi R, et al. Osteopontin is involved in the formation of malignant pleural effusion in lung cancer. Lung Cancer 2009;63:368-74. [Crossref] [PubMed]
- Heino J. Biology of tumor cell invasion: interplay of cell adhesion and matrix degradation. Int J Cancer 1996;65:717-22. [Crossref] [PubMed]
- Cheng D, Rodriguez RM, Perkett EA, et al. Vascular endothelial growth factor in pleural fluid. Chest 1999;116:760-5. [Crossref] [PubMed]
- Wang B, Shi L, Sun X, et al. Production of CCL20 from lung cancer cells induces the cell migration and proliferation through PI3K pathway. J Cell Mol Med 2016;20:920-9. [Crossref] [PubMed]
- Chen MC, Chen KC, Chang GC, et al. RAGE acts as an oncogenic role and promotes the metastasis of human lung cancer. Cell Death Dis 2020;11:265. [Crossref] [PubMed]
- Liu GN, Shi HZ, Xie ZH, et al. Epithelial neutrophil-activating peptide-78 recruits neutrophils into pleural effusion. Eur Respir J 2009;34:184-90. [Crossref] [PubMed]
- Neo SY, Lundqvist A. The Multifaceted Roles of CXCL9 Within the Tumor Microenvironment. Adv Exp Med Biol 2020;1231:45-51. [Crossref] [PubMed]
- Thielen C, Radermacher V, Trimeche M, et al. TARC and IL-5 expression correlates with tissue eosinophilia in peripheral T-cell lymphomas. Leuk Res 2008;32:1431-8. [Crossref] [PubMed]
- Yuan M, Zhu H, Xu J, et al. Tumor-Derived CXCL1 Promotes Lung Cancer Growth via Recruitment of Tumor-Associated Neutrophils. J Immunol Res 2016;2016:6530410. [Crossref] [PubMed]
- Qin XJ, Shi HZ, Huang ZX, et al. Interleukin-16 in tuberculous and malignant pleural effusions. Eur Respir J 2005;25:605-11. [Crossref] [PubMed]
- Ye LL, Peng WB, Niu YR, et al. Accumulation of TNFR2-expressing regulatory T cells in malignant pleural effusion of lung cancer patients is associated with poor prognosis. Ann Transl Med 2020;8:1647. [Crossref] [PubMed]
- Kaur S, Bronson SM, Pal-Nath D, et al. Functions of Thrombospondin-1 in the Tumor Microenvironment. Int J Mol Sci 2021;22:4570. [Crossref] [PubMed]
- Lunardi S, Lim SY, Muschel RJ, et al. IP-10/CXCL10 attracts regulatory T cells: Implication for pancreatic cancer. Oncoimmunology 2015;4:e1027473. [Crossref] [PubMed]
- Billottet C, Quemener C, Bikfalvi A. CXCR3, a double-edged sword in tumor progression and angiogenesis. Biochim Biophys Acta 2013;1836:287-95. [Crossref] [PubMed]
- Mulet M, Zamora C, Porcel JM, et al. Platelet factor 4 regulates T cell effector functions in malignant pleural effusions. Cancer Lett 2020;491:78-86. [Crossref] [PubMed]
- Aliper AM, Frieden-Korovkina VP, Buzdin A, et al. A role for G-CSF and GM-CSF in nonmyeloid cancers. Cancer Med 2014;3:737-46. [Crossref] [PubMed]
- Atanackovic D, Cao Y, Kim JW, et al. The local cytokine and chemokine milieu within malignant effusions. Tumour Biol 2008;29:93-104. [Crossref] [PubMed]
- Bradshaw M, Mansfield A, Peikert T. The role of vascular endothelial growth factor in the pathogenesis, diagnosis and treatment of malignant pleural effusion. Curr Oncol Rep 2013;15:207-16. [Crossref] [PubMed]
- Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev 2016;35:75-91. [Crossref] [PubMed]
- Jin HY, Lee KS, Jin SM, et al. Vascular endothelial growth factor correlates with matrix metalloproteinase-9 in the pleural effusion. Respir Med 2004;98:115-22. [Crossref] [PubMed]
- Yang K, Tian C, Zhang C, et al. The Controversial Role of IL-33 in Lung Cancer. Front Immunol 2022;13:897356. [Crossref] [PubMed]
- Wang H, Si S, Jiang M, et al. Leukemia inhibitory factor is involved in the pathogenesis of NSCLC through activation of the STAT3 signaling pathway. Oncol Lett 2021;22:663. [Crossref] [PubMed]
- Malekan M, Nezamabadi SS, Samami E, et al. BDNF and its signaling in cancer. J Cancer Res Clin Oncol 2023;149:2621-36. [Crossref] [PubMed]
- Han S, Wang T, Chen Y, et al. High CCL7 expression is associated with migration, invasion and bone metastasis of non-small cell lung cancer cells. Am J Transl Res 2019;11:442-52.
- Lv T, Zhao Y, Jiang X, et al. uPAR: An Essential Factor for Tumor Development. J Cancer 2021;12:7026-40. [Crossref] [PubMed]
- Fridlender ZG, Kapoor V, Buchlis G, et al. Monocyte chemoattractant protein-1 blockade inhibits lung cancer tumor growth by altering macrophage phenotype and activating CD8+ cells. Am J Respir Cell Mol Biol 2011;44:230-7. [Crossref] [PubMed]
- Deng J, Jiang R, Meng E, et al. CXCL5: A coachman to drive cancer progression. Front Oncol 2022;12:944494. [Crossref] [PubMed]
- Kuo PL, Huang MS, Hung JY, et al. Synergistic effect of lung tumor-associated dendritic cell-derived HB-EGF and CXCL5 on cancer progression. Int J Cancer 2014;135:96-108. [Crossref] [PubMed]
- Yu L, Guo Y, Chang Z, et al. Bidirectional Interaction Between Cancer Cells and Platelets Provides Potential Strategies for Cancer Therapies. Front Oncol 2021;11:764119. [Crossref] [PubMed]


