
Determinants of Aurora kinase B inhibitor sensitivity in small cell lung cancer
Introduction
There is considerable interest in targeting Aurora kinase B (AURKB) for small cell lung cancer (SCLC) treatment. Previous studies suggested the mutation statuses of tumor suppressors RB and p53, and the amplification status of MYC, alone could be used to identify SCLC tumors that would respond to AURKB inhibitors. A recent study by Ramkumar et al. challenges this notion and identifies BCL2 as a predictive marker and target to enhance AURKB inhibitor sensitivity in SCLC (1).
Background
SCLC accounts for ~15% of all lung cancer cases. This is an aggressive cancer sub-type with few treatment options and an exceedingly poor prognosis. The majority of SCLCs are diagnosed at an extensive stage (2). Standard systemic therapy for these patients involves platinum-based chemotherapy. Median survival times with this standard treatment are only 9–11 months with a 2-year survival rate of less than 5% (3). The introduction of immune checkpoint inhibitors has shown promise at improving SCLC patient survival in recent years (2). Nonetheless, the overall prognosis remains grim. Unlike non-SCLCs (NSCLCs) that in many cases can be treated with targeted agents, such as anti-EGFR compounds in EGFR-positive tumors, the availability of targeted agents for SCLC is limited. New targeted approaches are needed to improve outcomes in SCLC.
The Aurora kinases have emerged as therapy targets for SCLC. There are three aurora kinase family members: A, B, and C (4). AURKC expression is restricted to gametes where it regulates meiosis. In contrast, AURKA and AURKB are widely expressed and play critical roles in regulating mitosis. AURKA associates with the spindle poles where it regulates centrosome maturation and spindle assembly and entry into mitosis. In contrast, AURKB localizes at the kinetochore as part of the chromosome passenger complex where it does two things: first, it helps correct improper chromosome-spindle attachments as part of an error-correction process. Second, when spindles are detached from chromosomes, AURKB helps activate the spindle assembly checkpoint to block mitosis (5). Thus, inhibiting these kinases would be expected to disrupt central processes in mitosis and be toxic. High expressions of AURKA and/or AURKB are observed in multiple cancers and associated with poor patient outcomes, supporting that these kinases could be valuable targets for cancer treatment (6). A number of AURK inhibitors have been developed and tested in early clinical trials. However, most of these trials were halted due to bone marrow toxicity and poor clinical response. Some inhibitors targeted both A and B without specificity, and combined inhibition of both could contribute to at least some of the associated toxicity. AURKB seems a more promising drug target since AURKB inhibition causes mitotic slippage, catastrophic mitosis, and cell death, while AURKA inhibition is more associated with proliferation arrest (7). Thus, new inhibitors with heightened specificity, particularly for AURKB, have come to the fore in recent years. A number of clinical trials have been initiated with AURKB inhibitors, mostly for hematologic cancers but also some for solid tumors, including SCLC (6).
RB loss can render SCLCs sensitive to AURKB inhibitors
Certain molecular features of SCLCs may render them particularly sensitive to AURKB inhibitors. Nearly all (90–100%) SCLC tumors harbor loss-of-function alterations in the tumor suppressors RB and p53 (8). Approximately 20% of SCLCs have genomic amplification of a MYC-family oncogene (9). Oser and colleagues carried out a screen for genes in SCLC cells that when deleted caused toxicity specifically in cells that lacked RB, but not in cells with RB intact (10). The AURKB gene was identified in this screen, as were genes encoding proteins that interact with or function with AURKB, including components of the chromosome passenger complex. These findings suggested RB-deficient/mutated SCLC cells could be targeted by AURKB inhibitors, which they verified in cell lines and in xenograft and PDX tumor studies.
How might RB deficiency sensitize SCLC to AURKB inhibitors? RB-deficient cells have high levels of chromosome instability (CIN) (11). This is believed to be due to decreased H4K20 histone methylation in peri-centromeres which, in turn, decreases cohesin binding to chromosomes (11,12). The H4K20 methylation is mediated by SUV420H histone methyltransferase proteins. RB interacts with SUV420H, and the defects in histone methylation, cohesin binding, and CIN are reversed by overexpression of SUV420H in RB-deficient cells (11,13). Importantly, a recent study showed that AURKB is recruited to kinetochores when SUV420H is inhibited and, at kinetochores, helps prevent chromosome misalignments and missegregations (14). Lastly, RB loss increases activity of the transcription factor E2F1, which promotes AURKB gene expression (15). Thus, a model emerges in which RB loss decreases SUV420H activity and renders cells dependent on AURKB to maintain normal chromosome alignments (Figure 1A). Inhibition of AURKB in this RB-deficient setting results in toxic chromosome missegregations and cell death.
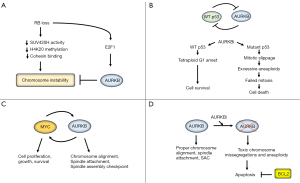
p53 deficient or mutated cells are hyper-sensitive to AURKB inhibitors
Studies by the Poon lab and others showed p53-deficiency renders cells especially sensitive to AURKB inhibition (7). How might p53 deficiency sensitize SCLC cells to AURKB inhibitors? p53 and AURKB function in a negative feedback loop that involves pRB-E2F1 and MDM2. Thus, E2F1 promotes AURKB gene expression (15). We showed that p53 represses expression of AURKB, AURKA, and other E2F1-responsive Aurora kinase pathway genes by promoting formation of transcription repressor complexes that include RB family proteins (so-called DREAM-complexes) (16). These DREAM complexes bind E2F1 and repress expression of E2F1 target genes. At the same time, AURKB is reported to phosphorylate p53, which increases p53 degradation by MDM2 (17). AURKB inhibition can cause chromosome misalignments and inhibit activation of the spindle assembly checkpoint, while also stabilizing and activating wild-type p53 (7). Activated p53 can then promote protective cell cycle arrests, including a tetraploid G1 arrest (4N arrest) in G2/M phase cells that blocks these cells from proceeding into mitosis with deficient spindle-chromosome attachments or misaligned chromosomes (18,19). In contrast, p53-mutant or deleted cells proceed with mitosis when AURKB is inhibited, followed by a failure of cytokinesis, mitotic slippage, and whole chromosome duplications that ultimately result in apoptosis and cell death when these hyperploid and aneuploid cells attempt to divide (7) (Figure 1B).
MYC amplification sensitizes SCLC cells to AURKB inhibitors
Amplification of MYC family genes (MYC, MYCL, MYCN) is observed in ~20% of SCLC tumors and associated with poor prognosis (9). Mollaoglu et al. established a mouse model of SCLC with transgenic expression of a stable MYC T58A mutant in a p53- and RB-deficient background (20). These mice developed SCLC that was metastatic and that, like the human condition, responded to initial chemotherapy treatment followed by rapid tumor recurrence. Importantly, these SCLC tumors and cells derived from them were sensitive to AURKA/B and AURKB-specific inhibitors alone and in combination with chemotherapy. How might MYC amplification render SCLCs sensitive to AURKB inhibition? MYC is an oncogenic transcription factor that promotes cell proliferation, growth, and survival. MYC-amplified cells, by virtue of being highly proliferative, may have a higher mitotic index and therefore be more sensitive to compounds like AURKB inhibitors that target mitotic cells. In addition, cells with MYC gene amplification are likely to depend, at least in part, on MYC protein to maintain proliferation and survival. Previous studies showed AURKB and MYC can function in a positive feedback loop where MYC promotes AURKB gene expression, and AURKB promotes MYC phosphorylation that stabilizes the MYC protein (21,22). It is unclear if this positive feedback loop is present in SCLC. Nonetheless, the findings suggest AURKB inhibition could target MYC-amplified cells in two ways: first, by inducing toxic chromosome mis-segregations and aneuploidy through its effect on AURKB in mitosis, and second by destabilizing and thus reducing the levels and activity of MYC (Figure 1C).
BCL2 is a predictive marker and target to enhance AURKB inhibitor sensitivity in SCLC
The results described above suggest the mutation/deletion status of RB and p53 genes, and/or the amplification status of MYC, could be used as a marker to identify SCLCs that would be sensitive to AURKB inhibition. Thus, the recent study from the Byers lab was particularly interesting (1). In their study, a large panel of 57 SCLC cell lines were tested for AURKB inhibitor sensitivity. The majority of cell lines were RB mutated/deleted and/or expressed high levels of MYC. Surprisingly, however, less than half of the cell lines displayed high or intermediate sensitivity to AURKB inhibitor, while the majority of cell lines were resistant. Some cell lines were resistant to AURKB inhibitor despite being RB-mutated/deleted or MYC-amplified, and some cell lines were sensitive to AURKB inhibition even in the absence of RB mutation/deletion or MYC amplification. The results indicated that RB and MYC status alone are imperfect predictors of AURKB inhibitor sensitivity. Next, the authors carried out an elegant proteomic RPPA analysis comparison between sensitive and resistant cell groups. This analysis revealed that AURKB inhibitor resistant cell lines expressed high levels of the anti-apoptotic protein BCL2. Silencing of BCL2 restored sensitivity in AURKB inhibitor-resistant cells, and BCL2 overexpression had the opposite effect, confirming that high BCL2 expression was the causal mediator of AURKB inhibitor resistance. Lastly, the authors showed that the BCL2 inhibitor venetoclax could sensitize BCL2-high expressing SCLC PDX tumors to AURKB inhibition in mice. Thus, high BCL2 expression can be a predictive marker of AURKB inhibitor resistance in SCLC that can be targeted by BCL2 inhibitors (Figure 1D).
Concluding remarks
Past clinical trials with AURKB inhibitors failed or were halted due to toxicity issues and poor clinical response in patients. At least three factors are likely to improve AURKB trials going forward. First is the inclusion of immune checkpoint inhibitors, which have already shown promise for improving outcomes in SCLC patients (2). At the time of this writing, there is at least one active clinical trial combining AURKB inhibitor with an immune checkpoint inhibitor in extensive-stage SCLC (NCT04745689). A second factor that may improve AURKB inhibitor trial success is improved drug formulations. The best example here is the AURKB inhibitor AZD2811. Nanoparticle formulations of AZD2811 have been shown to provide a slow but extended drug release that reduces toxicity while maintaining anti-tumor activity (23). A third factor that may improve AURKB inhibitor trial success is the development of predictive biomarkers of response. This is an on-going challenge. Past trials examined expressions of AURKA and B and also different proliferation and mitotic markers for their correlation with response and their potential as predictive markers (24). However, no consistent correlations were made and there are currently no established markers to differentiate responsive and non-responsive tumors. Recent studies suggested that the mutation/deletion status of RB and p53, and/or the amplification status of MYC, can be used to identify responsive tumors (7,10,20). The findings from Byers and colleagues suggest this is not necessarily the case. Specifically, their findings indicate BCL2 high vs. low expression can also identify responsive vs. non-responsive SCLC tumors (1). It is likely that combined analysis of RB/p53 status, MYC amplification, and BCL2 high vs. low expression status in tumors would best identify patients most likely to benefit from AURKB inhibitor treatment. Combining these analyses with gene expression patterns may also help stratify SCLC tumors into responsive and non-responsive groups even further (25). Related to toxicity, combination treatments that sensitize tumors to AURKB inhibition will allow lower treatment doses of AURKB inhibitor to be given and could therefore reduce toxicity. Results from the Byers lab study suggest the BCL2 inhibitor venetoclax can sensitize SCLC cells and tumors to the AURKB inhibitor AZD2811, which is in current clinical trials. Notably, venetoclax is already Food and Drug Administration (FDA) approved for AML, suggesting the potential for rapid clinical translation in SCLC for venetoclax in combination with AURKB inhibitors like AZD2811.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Lung Cancer Research. The article has undergone external peer review.
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-732/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-732/coif). L.D. received funding from the Swim Across America Foundation. C.G.M. received funding from the National Institutes of Health (NIH), the Department of Defense (DOD), the Bears Care Foundation, and the Swim Across America Foundation. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ramkumar K, Tanimoto A, Della Corte CM, et al. Targeting BCL2 Overcomes Resistance and Augments Response to Aurora Kinase B Inhibition by AZD2811 in Small Cell Lung Cancer. Clin Cancer Res 2023;29:3237-49. [Crossref] [PubMed]
- Saida Y, Watanabe S, Kikuchi T. Extensive-Stage Small-Cell Lung Cancer: Current Landscape and Future Prospects. Onco Targets Ther 2023;16:657-71. [Crossref] [PubMed]
- Kalemkerian GP, Loo BW, Akerley W, et al. NCCN Guidelines Insights: Small Cell Lung Cancer, Version 2.2018. J Natl Compr Canc Netw 2018;16:1171-82. [Crossref] [PubMed]
- Willems E, Dedobbeleer M, Digregorio M, et al. The functional diversity of Aurora kinases: a comprehensive review. Cell Div 2018;13:7. [Crossref] [PubMed]
- Krenn V, Musacchio A. The Aurora B Kinase in Chromosome Bi-Orientation and Spindle Checkpoint Signaling. Front Oncol 2015;5:225. [Crossref] [PubMed]
- Borah NA, Reddy MM. Aurora Kinase B Inhibition: A Potential Therapeutic Strategy for Cancer. Molecules 2021;26:1981. [Crossref] [PubMed]
- Marxer M, Ma HT, Man WY, et al. p53 deficiency enhances mitotic arrest and slippage induced by pharmacological inhibition of Aurora kinases. Oncogene 2014;33:3550-60. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Oser MG, Fonseca R, Chakraborty AA, et al. Cells Lacking the RB1 Tumor Suppressor Gene Are Hyperdependent on Aurora B Kinase for Survival. Cancer Discov 2019;9:230-47. [Crossref] [PubMed]
- Manning AL, Yazinski SA, Nicolay B, et al. Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol Cell 2014;53:993-1004. [Crossref] [PubMed]
- Hahn M, Dambacher S, Dulev S, et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev 2013;27:859-72. [Crossref] [PubMed]
- Gonzalo S, García-Cao M, Fraga MF, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 2005;7:420-8. [Crossref] [PubMed]
- Herlihy CP, Hahn S, Hermance NM, et al. Suv420 enrichment at the centromere limits Aurora B localization and function. J Cell Sci 2021;134:jcs249763. [Crossref] [PubMed]
- Kimura M, Uchida C, Takano Y, et al. Cell cycle-dependent regulation of the human aurora B promoter. Biochem Biophys Res Commun 2004;316:930-6. [Crossref] [PubMed]
- Duan L, Perez RE, Calhoun S, et al. RBL2/DREAM-mediated repression of the Aurora kinase A/B pathway determines therapy responsiveness and outcome in p53 WT NSCLC. Sci Rep 2022;12:1049. [Crossref] [PubMed]
- Gully CP, Velazquez-Torres G, Shin JH, et al. Aurora B kinase phosphorylates and instigates degradation of p53. Proc Natl Acad Sci U S A 2012;109:E1513-22. [Crossref] [PubMed]
- Shen H, Moran DM, Maki CG. Transient nutlin-3a treatment promotes endoreduplication and the generation of therapy-resistant tetraploid cells. Cancer Res 2008;68:8260-8. [Crossref] [PubMed]
- Davaadelger B, Shen H, Maki CG. Novel roles for p53 in the genesis and targeting of tetraploid cancer cells. PLoS One 2014;9:e110844. [Crossref] [PubMed]
- Mollaoglu G, Guthrie MR, Böhm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- den Hollander J, Rimpi S, Doherty JR, et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 2010;116:1498-505. [Crossref] [PubMed]
- Jiang J, Wang J, Yue M, et al. Direct Phosphorylation and Stabilization of MYC by Aurora B Kinase Promote T-cell Leukemogenesis. Cancer Cell 2020;37:200-215.e5. [Crossref] [PubMed]
- Ashton S, Song YH, Nolan J, et al. Aurora kinase inhibitor nanoparticles target tumors with favorable therapeutic index in vivo. Sci Transl Med 2016;8:325ra17. [Crossref] [PubMed]
- Falchook GS, Bastida CC, Kurzrock R. Aurora Kinase Inhibitors in Oncology Clinical Trials: Current State of the Progress. Semin Oncol 2015;42:832-48. [Crossref] [PubMed]
- Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346-360.e7. [Crossref] [PubMed]


