
Oncogene alterations in non-small cell lung cancer with FGFR1 amplification—novel approach to stratify patients who benefit from FGFR inhibitors
Lung cancer is the principal cause of cancer-related deaths worldwide (1). The discovery of driver oncogenes in non-small cell lung cancer (NSCLC) has revolutionized treatment for this disease. Therapies to driver alterations such as mutations in EGFR, BRAF, and ROS1, and translocation of the ALK, are now available in clinical practice. The appropriate implementation of targeted therapies has significantly prolonged the survival of NSCLC patients with driver oncogenes (2,3). In contrast to the significant progress achieved in targeted therapy for lung adenocarcinoma, the identification and targeting of driver mutations in squamous cell lung cancer (SQCLC) remains to be established.
One potential molecular target in SQCLC is the fibroblast growth factor receptor 1 (FGFR1). In SQCLC, FGFR1 amplification is detected in approximately 13–22% of patients (4-6). The FGFR pathway plays critical roles in cell migration, mitosis, and death, suggesting a role for this network in the oncogenic process (7). Deregulation of FGFR signaling has been reported in a variety of human cancers such as breast and bladder, and FGFR inhibitors have been approved by the U.S. Food and Drug Administration (FDA) for the treatment of FGFR2-fusion cholangiocarcinoma and FGFR3-mutated urothelial carcinoma (8). In contrast to the demonstrated efficacy of these agents in targeting FGFR fusions/mutations in other types of cancers, FGFR inhibitors show only a modest response in FGFR1 amplified SQCLC (9-11). As such, there is a clear need to elucidate the mechanisms underlying the heterogeneous response of SQCLC to FGFR inhibitors, and to establish markers that predict response to these agents.
The driver role of FGFR1 amplification could be affected by neighboring genes in the 8p11–12 locus (12). The 8p12 region includes multiple centers of amplification with chromosomal breakpoints spanning an unusually large region surrounding the peak (13). Therefore, FGFR1 amplification is associated with many other genes located in the 8p11–12 locus, such as BAG4, NSD3 (WHSC1L1), LSM1, ASH2L, DDHD2, and EIF4EBP1. In one study, only 28% of cases with 8p12 amplification had peaks that centered on FGFR1, raising the possibility that FGFR1 is not the driver in the majority of cases with 8p12 amplification (13). In line with this, other genes located in the 8p11–12 locus were also reported to contribute to oncogenesis (14). For example, NSD3 was identified as an alternative driver in FGFR1-amplified tumors (15). NSD3 is a lysine methyltransferase involved in various cellular processes, including transcriptional regulation and DNA damage repair (16-18). Yuan et al. identified that suppression of NSD3, but not of FGFR1, diminished tumor growth and prolonged survival in a mouse model of SQCLC (19). They also showed that knockout of NSD3 in patient-derived xenografts from primary SQCLC with NSD3 amplification attenuated tumor growth in vivo.
In addition to its genetic heterogeneity, the expression of FGFR1 messenger RNA (mRNA) is not well correlated with the magnitude of FGFR1 gene amplification. Kotani et al. showed that there is a discrepancy between the genetic amplification and protein expression of FGFR1 in FGFR1-amplified SQCLC cell lines. While cell lines with high FGFR1 amplification and protein expression are sensitive to FGFR inhibitor monotherapy, activation of additional driver oncogenes such as HER2 and PDGFR mitigate the effect of FGFR1 inhibition in FGFR1 amplified but low protein-expressing cells (20). In addition, among 25 patients with FGFR1 amplified SQCLC identified by fluorescence in situ hybridization (FISH) analysis, only six tumors were positive for FGFR1 protein expression. Similarly, Wynes et al. demonstrated that sensitivity to the FGFR inhibitor ponatinib [half-maximal inhibitory concentration (IC50) value ≤50 nM] was correlated with FGFR1 mRNA and protein expression, but not FGFR1 gene copy number in lung cancer cell lines (21). Consistent with preclinical findings, FGFR1 protein expression was only detected in 35.5% of FGFR1 amplified cases, while FGFR1 protein expression was also detected in 29.6% of non-amplified cases among 208 SQCLC tumor samples (22). Likewise, in a phase II study with the FGFR inhibitor dovitinib, among 16 patient samples with FGFR1 amplification, 31.2% showed medium/high expression, while 43.7% showed low expression, and 25% showed no FGFR1 mRNA expression (11). Furthermore, this study did not demonstrate a correlation between FGFR1–3 mRNA level and response to dovitinib, although the sample size was limited. These data indicate that FGFR1 amplification is not indicative of its mRNA or protein expression, and even high FGFR1 protein expression may not be enough to explain the efficacy to FGFR inhibitors.
Recently, Malchers et al. demonstrated that there’s a correlation between the oncogenic ectodomain deletion of FGFR1 caused by somatic rearrangements with tumor dependency on FGFR1 and sensitivity to FGFR inhibitors in SQCLC (23). By performing deep genomic sequencing of SQCLC samples with FGFR1 amplification, including patient samples treated with FGFR inhibitors, they identified two types of tail-to-tail rearrangements that happen within or close to the FGFR1 gene (Figure 1).
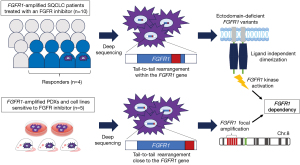
Tail-to-tail rearrangements within the FGFR1 gene were identified in 8% of FGFR1 amplified samples. Although tail-to-tail rearrangements occur within the open reading frame, the truncated FGFR1 protein can still be transcribed using a noncanonical in-frame ATG start codon, resulting in various deletions in the extracellular region of the protein. The loss of ectodomain components causing FGFR1 dependence is presumably mediated by the enhancement of ligand-independent dimerization, leading to sensitivity to FGFR inhibitors. Interestingly, half of the patients who responded to FGFR inhibitors harbored FGFR1 amplification due to tail-to-tail rearrangements within the FGFR1 gene, while these rearrangements were not detected in non-responders.
Tail-to-tail rearrangements were also found in the locus that is close to the FGFR1 gene. These rearrangements led to an FGFR1-centered amplification pattern, and they frequently co-existed with disruptive rearrangements of NSD3. Tail-to-tail rearrangements close to FGFR1 with NSD3-disruptive rearrangements were found in the NCI-H1581 and DMS114 cell lines, both of which are well known to be sensitive to FGFR inhibitors. Furthermore, tail-to-tail rearrangements in or close to the FGFR1 gene were detected in seven of nine cases in a collective analysis of cell lines, patient-derived xenograft models, and patient-derived specimens that were sensitive to FGFR1 inhibitors.
Mechanistically, 8p11 amplification and FGFR1 dependence were at least partly mediated by break-fusion-bridging (BFB), a mechanism that leads to complex genome rearrangements in multiple cancers (24). A BFB cycle begins with chromosomal rearrangements mediated by the foldback inversion of two sister chromatids due to a lack of telomeres during DNA replication, resulting in two centromeres in the fused bridge. These foldback inversions were mediated by formation of single-stranded DNA hairpins. In the anaphase, two centromeres are stretched to opposite poles. Then, the two sister chromatids are split with a double-strand break on the bridge between the two centromeres. Since each daughter cell contains chromatids without telomeres, the BFB cycle can repeat. Repetition of BFB cycles contributes to a surge in stair-like copy number amplifications and foldback inversions. Given that BFB can affect a number of genes besides FGFR1, and tail-to-tail rearrangements can be induced by other mechanisms such as chromothripsis, these tail-to-tail rearrangements randomly happen, and their occurrence within or close to the FGFR1 gene likely promote a survival advantage.
The study by Malchers et al. (23) suggests that the identification of tail-to-tail rearrangements within or close to the FGFR1 gene could be a potential new approach to select patients for FGFR inhibitor treatment. However, FGFR1 amplified SQCLC is still heterogenous. While two of the four patients who responded to FGFR1 inhibitors did not show tail-to-tail rearrangements within the FGFR1 gene, rearrangements within or close to FGFR1 were also observed in approximately 25% of the FGFR1 inhibitor-insensitive models. First, it would be important to know how much FGFR1 mRNA and protein is expressed in tumors harboring tail-to-tail rearrangements, as discrepancies between gene amplification and mRNA expression of FGFR1 have been reported. Second, the real value of FGFR1 amplification remains to be fully elucidated. In the case of tail-to-tail rearrangements within the FGFR1 gene, the ectodomain-lacking FGFR1 caused FGFR1 oncogene dependency in Ba/F3 cells. In contrast, although tail-to-tail rearrangements upstream of FGFR1 lead to FGFR1-centered amplification in the 8p11 locus, it is unclear whether its amplification is essential for tumor cell survival. Furthermore, tumors with tail-to-tail rearrangements close to FGFR1 were accompanied by destructive rearrangements in NSD3. This phenomenon can potentially induce unfavorable conditions for tumor growth, given the proliferative role of NSD3 (19).
A series of studies by the authors illustrates the complexity of SQCLC (13,23,25), probably due to tobacco smoking. It is hoped that continued research efforts will eventually build a targeted strategy for this difficult population.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Lung Cancer Research. The article has undergone external peer review.
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-777/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-777/coif). H.E. reports honoraria from AMGEN, AstraZeneca, Bristol-Myers Squibb, Chugai Pharmaceutical, Incyte, Kyowa-Kirin, Merck Serono, Takeda, and Taiho. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Araghi M, Mannani R, Heidarnejad Maleki A, et al. Recent advances in non-small cell lung cancer targeted therapy; an update review. Cancer Cell Int 2023;23:162. [Crossref] [PubMed]
- Tan AC, Tan DSW. Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J Clin Oncol 2022;40:611-25. [Crossref] [PubMed]
- Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. [Crossref] [PubMed]
- Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 2013;31:731-7. [Crossref] [PubMed]
- Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol 2012;7:1775-80. [Crossref] [PubMed]
- Xie Y, Su N, Yang J, et al. FGF/FGFR signaling in health and disease. Signal Transduct Target Ther 2020;5:181. [Crossref] [PubMed]
- Krook MA, Reeser JW, Ernst G, et al. Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br J Cancer 2021;124:880-92. [Crossref] [PubMed]
- Aggarwal C, Redman MW, Lara PN Jr, et al. SWOG S1400D (NCT02965378), a Phase II Study of the Fibroblast Growth Factor Receptor Inhibitor AZD4547 in Previously Treated Patients With Fibroblast Growth Factor Pathway-Activated Stage IV Squamous Cell Lung Cancer (Lung-MAP Substudy). J Thorac Oncol 2019;14:1847-52. [Crossref] [PubMed]
- Nogova L, Sequist LV, Perez Garcia JM, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol 2017;35:157-65. [Crossref] [PubMed]
- Lim SH, Sun JM, Choi YL, et al. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: A single-arm, phase 2 study. Cancer 2016;122:3024-31. [Crossref] [PubMed]
- Voutsadakis IA. Amplification of 8p11.23 in cancers and the role of amplicon genes. Life Sci 2021;264:118729. [Crossref] [PubMed]
- Malchers F, Dietlein F, Schöttle J, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov 2014;4:246-57. [Crossref] [PubMed]
- Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011;6:e20351. [Crossref] [PubMed]
- Satpathy S, Krug K, Jean Beltran PM, et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell 2021;184:4348-4371.e40. [Crossref] [PubMed]
- Rooney C, Geh C, Williams V, et al. Characterization of FGFR1 Locus in sqNSCLC Reveals a Broad and Heterogeneous Amplicon. PLoS One 2016;11:e0149628. [Crossref] [PubMed]
- Xiu S, Chi X, Jia Z, et al. NSD3: Advances in cancer therapeutic potential and inhibitors research. Eur J Med Chem 2023;256:115440. [Crossref] [PubMed]
- Li D, Tian T, Ko CN, et al. Prospect of targeting lysine methyltransferase NSD3 for tumor therapy. Pharmacol Res 2023;194:106839. [Crossref] [PubMed]
- Yuan G, Flores NM, Hausmann S, et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 2021;590:504-8. [Crossref] [PubMed]
- Kotani H, Ebi H, Kitai H, et al. Co-active receptor tyrosine kinases mitigate the effect of FGFR inhibitors in FGFR1-amplified lung cancers with low FGFR1 protein expression. Oncogene 2016;35:3587-97. [Crossref] [PubMed]
- Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res 2014;20:3299-309. [Crossref] [PubMed]
- Elakad O, Lois AM, Schmitz K, et al. Fibroblast growth factor receptor 1 gene amplification and protein expression in human lung cancer. Cancer Med 2020;9:3574-83. [Crossref] [PubMed]
- Malchers F, Nogova L, van Attekum MH, et al. Somatic rearrangements causing oncogenic ectodomain deletions of FGFR1 in squamous cell lung cancer. J Clin Invest 2023;133:e170217. [Crossref] [PubMed]
- Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010;467:1109-13. [Crossref] [PubMed]
- Malchers F, Ercanoglu M, Schütte D, et al. Mechanisms of Primary Drug Resistance in FGFR1-Amplified Lung Cancer. Clin Cancer Res 2017;23:5527-36. [Crossref] [PubMed]


