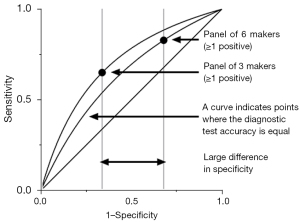
Improved diagnostic accuracy with three lung tumor markers compared to six-marker panel
Highlight box
Key findings
• We retrospectively reviewed 1,733 patients with suspected lung cancer using tumor markers as index tests and clinical diagnosis by the physician as the reference index; the combination of three tumor markers, namely carcinoembryonic antigen (CEA), cytokeratin-19 fragment (CYFRA), and neuron-specific enolase (NSE) provided better diagnostic test accuracy than panel of six tumor markers, especially regarding false positive rate.
What is known and what is new?
• Some liked to check as many as tumor markers in fear of missing lung cancer patients. Single tumor marker does not have sufficient diagnostic test accuracy.
• We showed that the panel of three markers had better diagnostic value than that of six. CEA, CYFRA, and NSE had reasonable diagnostic test accuracy.
What is the implication, and what should change now?
• We would like to recommend to check the panel of three tumor markers for patients with suspected lung cancer.
Introduction
Lung cancer remains the leading cause of cancer death, with 2.2 million new cases and approximately 1.8 million deaths reported worldwide (1). Lung cancer is divided into small-cell lung cancer and non-small-cell lung cancer. Diagnosing lung cancer is crucial for timely and appropriate treatment and better patient outcomes for both pathological types. Serum tumor markers such as carcinoembryonic antigen (CEA), cytokeratin-19 fragment (CYFRA), squamous cell carcinoma-associated antigen (SCC), neuron-specific enolase (NSE), pro-gastrin-releasing peptide (ProGRP), and sialyl Lewis-X antigen (SLX) are often measured in patients with suspected lung cancer. This is because serum tumor marker measurements based on blood tests are minimally invasive and widely available in most of medical facilities. A known limitation of tumor markers in diagnosing lung cancer is the accuracy of the diagnostic tests. To improve the sensitivity, physicians are likely to check multiple tumor markers. For example, Molina et al. recommended checking six tumor markers (2).
However, measuring multiple tumor markers (≥1 abnormal tumor marker value) increases the false positive (2). False positive tumor markers for diagnosing purposes may lead to unnecessary invasive examinations such as trans-bronchoscopic lung biopsy and video-assisted thoracic surgery. Moreover, the tumor marker panel of compound 6 may be expensive, particularly in developing countries. Therefore, we aimed to determine the minimal necessary combination of tumor markers. In this study, we examined the performance of individual and combined serum tumor markers for diagnosing lung cancer. This manuscript is written following STARD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-855/rc).
Methods
Study overview
This was a single-center, retrospective, and observational study. This study was reviewed and approved by the Ethics Committee for Clinical Research of the Yokohama City University Hospital (approval No. F220800003). This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The study was designed in compliance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects by Japanese Ministry of Health, Labour and Welfare. This guideline does not request informed consent for a retrospective review but does demand that researchers provide subjects with an opportunity to opt out. We issued an opt-out notice; however, there were no patients who wished to opt out.
Patients
We reviewed the charts of patients with suspected lung cancer who underwent all six tumor marker tests (CEA, CYFRA, SCC, NSE, ProGRP, and SLX) on the same day between April 2001 and May 2022 at the Department of Respiratory Medicine, Yokohama City University Hospital. Patients missing at least one of the six tumor markers were excluded because the multi-variable analysis was not applicable.
Lung cancer diagnosis
Patients confirmed to have lung cancer based on International Classification of Diseases, 10th revision (ICD-10) were regarded as having lung cancer. Physicians can provide the ICD-10 code based on clinical judgement without pathological confirmation. They might know the results of tumor markers.
Tumor markers
Serum CEA, SCC, and CYFRA values were measured in the hospital within 24 h of sampling without knowing physicians’ diagnosis; SLX, ProGRP, and NSE were measured by an outside laboratory. CEA levels were measured by an electrochemiluminescence immunoassay using the Eclusis reagent CEA II (Roche Diagnostics, Tokyo, Japan). SCC antigen levels were measured by a chemiluminescence immunoassay using Alinity SCC Abbott or ARCHITECT SCC (Abbott Japan, Tokyo, Japan). Chemiluminescence immunoassay using Alinity SCC Abbott or ARCHITECT SCC (Abbott Japan) was used to detect CYFRA. SLX was assessed by radioimmunoassay (RIA) solid phase method using SLX “DP” IRMA Kit (DENIS Pharma Co., Tokyo, Japan). A chemiluminescent enzyme immunoassay, Lumipulse Presto ProGRP (Fujirebio Co., Tokyo, Japan), was used to evaluate ProGRP levels. NSE levels were measured by electrochemiluminescence immunoassay using Eclucis reagent NSE (Roche Diagnostics). When the tumor marker level was too high for each assay, the blood sample was diluted using the appropriate reagent, and there was no upper limit to the measurement.
The lower limit of tumor marker detection was set as follows, but none of the data in this study fell below these levels: CEA (0.3 ng/mL), SCC (0.5 ng/mL), CYFRA (0.1 ng/mL), SLX (3.5 U/mL), ProGRP (2.0 pg/mL), NSE (0. 075 ng/mL). The upper limits of normal for each of the six tumor markers were determined based on published articles: CEA 5 ng/mL, CYFRA 3.3 ng/mL, SCC 2 ng/mL, NSE 25 ng/mL, ProGRP 50 pg/mL, and SLX 38.0 U/mL (3,4). Due to the nature of tumor markers, there was no indeterminant result.
Statistical analysis
First, we evaluated the diagnostic accuracy of the uncombined tumor markers. The area under the receiver operating characteristic (ROC) curve (AUC) was determined for each of the six markers. For this analysis, the raw values of the markers and binary variables (> the upper limit of normal) were applied. Second, we proposed a new panel with fewer markers. All six markers were used as explanatory variables in a logistic regression model to predict confirmed lung cancer. We composited two models using either common logarithmically transformed or binarized (> the upper limit of normal) markers. Using a logistic regression model, we designed a panel of tumor markers that had a significant impact on lung cancer diagnosis. Third, the AUC of the newly suggested panel and the panel of the six markers were compared.
All statistical analyses were performed using the GraphPad Prism 9 software (GraphPad Software, Inc., San Diego, CA, USA). Statistical significance was set at P<0.05. In the logistic regression analysis, P values for each explanatory variable were multiplied by six as a Bonferroni correction.
Results
Characterization of participants
The backgrounds of the 1,733 included patients are shown in Table 1. The median age of the population was 72 years, including 1,128 males (65.1%) and 605 females (34.9%). Median and interquartile range (25–75 percentiles) of tumor marker values are shown in Table 1: CEA 3.2 (1.9–5.6) ng/mL, CYFRA 2.4 (1.6–4.1) ng/mL, SCC 1.1 (0.7–1.6) ng/mL, NSE 10.9 (8.9–13.1) ng/mL, ProGRP was 45.6 (34.4–62.9) pg/mL, and SLX 32 (27–39.5) U/mL. The number of patients with tumor markers above the upper limit of normal was 504 (29.1%) for CEA, 575 (33.2%) for CYFRA, 273 (15.8%) for SCC, 86 (5.0%) for NSE, 733 (42.3%) for ProGRP, and 461 (26.6%) for SLX. Of the 1,733 patients, 779 (45%) had a confirmed diagnosis of lung cancer.
Table 1
| Characteristics | Value (n=1,733) |
|---|---|
| Age (years) | 72 [63–78] |
| Gender | |
| Male | 1,128 (65.1) |
| Female | 605 (34.9) |
| Raw value of tumor markers | |
| CEA (ng/mL) | 3.2 [1.9–5.6] |
| CYFRA (ng/mL) | 2.4 [1.6–4.1] |
| SCC (ng/mL) | 1.1 [0.7–1.6] |
| NSE (ng/mL) | 10.9 [8.9–13.1] |
| ProGRP (pg/mL) | 45.6 [34.4–62.9] |
| SLX (U/mL) | 32 [27–39.5] |
| Exceeding upper limit | |
| CEA (5.0 ng/mL) | 504 (29.1) |
| CYFRA (3.3 ng/mL) | 575 (33.2) |
| SCC (2.0 ng/mL) | 273 (15.8) |
| NSE (25.0 ng/mL) | 86 (5.0) |
| ProGRP (50.0 pg/mL) | 733 (42.3) |
| SLX (38.0 U/mL) | 461 (26.6) |
| Lung cancer | |
| Yes | 779 (45.0) |
| No | 954 (55.0) |
Data are presented as median [range] or n (%). CEA, carcinoembryonic antigen; CYFRA, cytokeratin-19 fragment; SCC, squamous cell carcinoma-associated antigen; NSE, neuron-specific enolase; ProGRP, pro-gastrin-releasing peptide; SLX, sialyl Lewis X antigen.
Diagnostic performance of single tumor marker
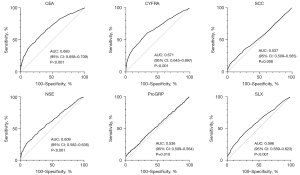
ROC curves were drawn for each raw tumor marker to predict confirmed cancer diagnosis (Figure 1). The tumor marker with the highest diagnostic accuracy was CEA [AUC =0.683, 95% confidence interval (CI): 0.658–0.709], followed by CYFRA (AUC =0.671, 95% CI: 0.645–0.697), NSE (AUC =0.609, 95% CI: 0.582–0.636) and SLX (AUC =0.596, 95% CI: 0.569–0.623). SCC (AUC =0.537, 95% CI: 0.509–0.565) and ProGRP (AUC =0.536, 95% CI: 0.509–0.564) had poorer AUC.
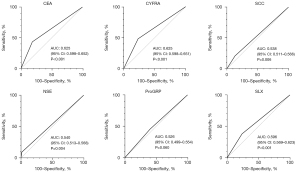
Diagnostic test accuracy of each of the tumor markers as binary variables (≥ upper limit of normal) for confirmed lung cancer was evaluated as shown in Figure 2. We found good AUC for CEA (AUC =0.625, 95% CI: 0.598–0.652), CYFRA (AUC =0.625, 95% CI: 0.598–0.651), and SLX (AUC =0.596, 95% CI: 0.569–0.623), whereas the AUC of SCC, NSE, and ProGRP were poor.
New panel of fewer markers
To establish a reasonable combination of tumor markers, logistic regression analysis was performed to predict confirmed lung cancer. In a model of logarithmic transformed levels, three markers were significantly associated with the confirmed lung cancer diagnosis (upper part of Table 2): CEA [odds ratio (OR) 3.23, 95% CI: 2.43–4.32], CYFRA (OR 3.04, 95% CI: 2.10–4.44), and NSE (OR 3.88, 95% CI: 1.99–7.69). Even after the tumor marker levels were binarized, the same three markers were significantly related to lung cancer: CEA (OR 2.40, 95% CI: 1.90–3.04), CYFRA (OR 2.02, 95% CI: 1.60–2.54), and NSE (OR 4.18, 95% CI: 2.34–7.96).
Table 2
| Tumor markers | Odds ratio (95% CI) or odds ratio |
P value | Pc value |
|---|---|---|---|
| Logarithmic transformed model | |||
| CEA | 3.23 (2.43–4.32) | <0.001 | <0.001 |
| CYFRA | 3.04 (2.10–4.44) | <0.001 | <0.001 |
| SCC | 1.11 (0.77–1.60) | 0.5904 | 0.999 |
| NSE | 3.88 (1.99–7.69) | <0.001 | <0.001 |
| ProGRP | 1.02 (0.69–1.52) | 0.9204 | 0.999 |
| SLX | 1.10 (0.59–2.06) | 0.7856 | 0.999 |
| Binary model | |||
| CEA | 2.40 | <0.001 | <0.001 |
| CYFRA | 2.02 | <0.001 | <0.001 |
| SCC | 1.19 | 0.2212 | 0.999 |
| NSE | 4.18 | <0.001 | <0.001 |
| ProGRP | 0.97 | 0.7914 | 0.999 |
| SLX | 1.34 | 0.0111 | 0.067 |
For the logarithmic transformed model, tumor marker values were log-transformed before logistic regression analysis. Therefore, the odds ratio was expressed as every 10-fold increase in tumor markers. For the binary model, any tumor marker that exceeded the upper normal limit was counted. Pc value, corrected P value (Bonferroni correction); CI, confidence interval; CEA, carcinoembryonic antigen; CYFRA, cytokeratin-19 fragment; SCC, squamous cell carcinoma-associated antigen; NSE, neuron-specific enolase; ProGRP, pro-gastrin-releasing peptide; SLX, sialyl Lewis X antigen.
Diagnostic performance of the all six markers and the new panel of three
Finally, the AUC of the new panel (CEA, CYFRA, and NSE) and the panel that included all six markers were compared (Figure 3). For this analysis, patients with at least one marker above the upper limit of normal were considered positive.
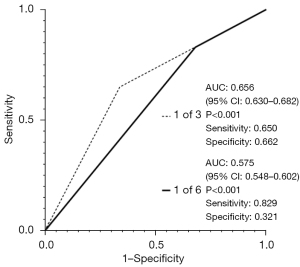
The combination of three tumor markers (AUC =0.656, 95% CI: 0.630–0.682, sensitivity 0.650, specificity 0.662) had better diagnostic performance than six tumor markers (AUC =0.575, 95% CI: 0.548–0.602, sensitivity 0.829, specificity 0.321). Notably, the measurement of the six tumor markers led to increased sensitivity and decreased specificity (Figure 4).
Discussion
In this study, we found that the combined assessment of the three serum tumor markers, CEA, CYFRA21-1, and NSE (one or more abnormal values), resulted in a better lung cancer diagnosis than the assessment of all six tumor markers (Figure 3). Simultaneous assessment of the six tumor markers (one or more abnormal values) resulted in more frequent false positives.
This study showed that, among the six markers, CEA had the best diagnostic accuracy for lung cancer. CEA is the most well-known tumor marker of epithelial malignancies. CEA has been used to diagnose lung cancer for half a century since an early study reported higher mean serum CEA levels in 487 lung cancer patients than in 228 healthy blood donors (5). Later, CEA levels were known to elevate, especially in lung adenocarcinoma. However, no studies have shown that CEA alone is useful for diagnosing lung cancer.
Molina et al. studied lung cancer serum tumor markers in 211 patients with non-small-cell lung cancer and found that CEA and CA125 were significantly higher in adenocarcinoma, and CEA <10 ng/mL could exclude adenocarcinoma in 82% of cases (6). CEA increases with age, smoking, inflammatory bowel disease, pancreatitis, cirrhosis, renal dysfunction, and hypothyroidism, which may cause false positive CEA without malignancy (7-9). High blood glucose, severe arterial stiffness, and increased visceral fat area also raise CEA levels (10-12).
CYFRA21-1 is specifically expressed in the epithelial tissues of the small intestine, colon, liver lactotrophs, pancreas, gallbladder, bladder, and bronchus (13). Renal dysfunction is an ill-reputed factor that causes false-positive CYFRA21-1. Nakahama et al. measured serum CYFRA21-1 levels in patients with renal failure and reported that CYFRA21-1 was elevated in 21% of patients undergoing hemodialysis (14). Elevated CYFRA21-1 levels have been reported in other benign lung diseases, such as chronic obstructive pulmonary disease (COPD) (15).
NSE is localized to neurons in mammalian nervous tissue and is also present in peripheral and central neuroendocrine cells (16). Therefore, NSE is often elevated in diseases associated with neuronal damage. Several studies have shown that NSE is useful in diagnosing and monitoring small-cell carcinoma of the lung (17-19). Besides small-cell carcinoma of the lung, NSE is also a tumor marker for melanoma, seminoma, carcinoid tumor, Merkel cell tumor, immature teratoma, malignant pheochromocytoma, and other neuroendocrine tumors (20).
Using combined panel of six tumor markers for lung cancer can lead to overdiagnosis, overtreatment, and overutilization due to the high rate of false-positive results. Normal values for tumor markers are often set to include 95% of healthy individuals. A single tumor marker can produce a 5% false positive rate, and testing six tumor markers can nearly increase the false positive rate to 30%. This can result in unnecessary invasive tests and interventions with risks and side effects. Over-intervention refers to unnecessary bronchoscopy and surgical biopsy despite the absence of significant disease progression or clinical benefit. Overutilization occurs when tumor markers are excessively used without considering their limitations, leading to unnecessary diagnostic procedures, invasive tests, and increased healthcare costs. False-positive results from tumor marker testing can trigger anxiety and distress in patients, prompting additional investigations and interventions that may not be justified. Addressing these challenges requires the development and implementation of diagnostic methods that enhance accuracy, reduce false positives, facilitate appropriate clinical decision-making, minimize patient harm, and optimize resource allocation in the management of lung cancer.
In the diagnosis of lung cancer, histopathological and cytopathological examinations through methods such as bronchoscopy, video-associated thoracic surgery, computed tomography (CT)-guided lung biopsy, and thoracentesis have been the standard. However, in the real world, it is not always possible to obtain pathological specimens, necessitating the exploration of lung cancer possibilities through alternative methods. Serum tumor markers have been one of the traditional alternatives, but recent years have seen a growing interest in new diagnostic approaches that accommodate a variety of specimens, including liquid biopsies, facilitated by next-generation sequencing and new genetic biomarkers (21,22).
A limitation of this study is that data for the histopathological type and stage were not available. Another limitation is possible bias owing to a single-center study design.
Conclusions
We examined the performance of tumor markers in lung cancer diagnosis using data from 1,733 patients. Compared to applying all six markers (at least one marker above the upper limit of normal), the panel with three markers (at least one marker above the upper limit of normal) led to a better predictive value by lowering the risk of false positives at the cost of slightly dismissing sensitivity.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-855/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-855/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-855/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-855/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was reviewed and approved by the Ethics Committee for Clinical Research of the Yokohama City University Hospital (approval No. F220800003). This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was designed in compliance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects by Japanese Ministry of Health, Labour and Welfare. This guideline does not request informed consent for a retrospective review but does demand that researchers provide subjects with an opportunity to opt out. We issued an opt-out notice; however, there were no patients who wished to opt out.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Molina R, Marrades RM, Augé JM, et al. Assessment of a Combined Panel of Six Serum Tumor Markers for Lung Cancer. Am J Respir Crit Care Med 2016;193:427-37. [Crossref] [PubMed]
- Pavićević R, Milicić J, Bubanović G, et al. Serum tumor marker CYFRA 21-1 in the diagnostics of NSCLC lung cancer. Coll Antropol 1998;22:629-35.
- Molina R, Augé JM, Bosch X, et al. Usefulness of serum tumor markers, including progastrin-releasing peptide, in patients with lung cancer: correlation with histology. Tumour Biol 2009;30:121-9. [Crossref] [PubMed]
- Vincent RG, Chu TM, Fergen TB, et al. Carcinoembryonic antigen in 228 patients with carcinoma of the lung. Cancer 1975;36:2069-76. [Crossref] [PubMed]
- Molina R, Filella X, Augé JM, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol 2003;24:209-18. [Crossref] [PubMed]
- Witherspoon LR, Shuler SE, Alyea K, et al. Carcinoembryonic antigen: assay following heat compared with perchloric acid extraction in patients with colon cancer, non-neoplastic gastrointestinal diseases, or chronic renal failure. J Nucl Med 1983;24:916-21.
- Amino N, Kuro R, Yabu Y, et al. Elevated levels of circulating carcinoembryonic antigen in hypothyroidism. J Clin Endocrinol Metab 1981;52:457-62. [Crossref] [PubMed]
- Khoo SK, Mackay IR. Carcinoembryonic antigen in serum in diseases of the liver and pancreas. J Clin Pathol 1973;26:470-5. [Crossref] [PubMed]
- Lee JY, Lee HK, Lee DC, et al. Serum carcinoembryonic antigen is associated with abdominal visceral fat accumulation in female Korean nonsmokers. PLoS One 2012;7:e43518. [Crossref] [PubMed]
- Bae U, Shim JY, Lee HR, et al. Serum carcinoembryonic antigen level is associated with arterial stiffness in healthy Korean adult. Clin Chim Acta 2013;415:286-9. [Crossref] [PubMed]
- No JI, Yang JY, Hyun HJ, et al. Factors Associated with Serum Levels of Carcinoembryonic Antigen in Healthy Non-smokers. Korean J Fam Med 2013;34:413-9. [Crossref] [PubMed]
- Omary MB, Ku NO, Strnad P, et al. Toward unraveling the complexity of simple epithelial keratins in human disease. J Clin Invest 2009;119:1794-805. [Crossref] [PubMed]
- Nakahama H, Tanaka Y, Fujita Y, et al. CYFRA 21-1 and ProGRP, tumor markers of lung cancer, are elevated in chronic renal failure patients. Respirology 1998;3:207-10. [Crossref] [PubMed]
- Xu RH, Liao CZ, Luo Y, et al. Optimal cut-off values for CYFRA 21-1 expression in NSCLC patients depend on the presence of benign pulmonary diseases. Clin Chim Acta 2015;440:188-92. [Crossref] [PubMed]
- Schmechel D, Marangos PJ, Brightman M. Neurone-specific enolase is a molecular marker for peripheral and central neuroendocrine cells. Nature 1978;276:834-6. [Crossref]
- Akoun GM, Scarna HM, Milleron BJ, et al. Serum neuron-specific enolase. A marker for disease extent and response to therapy for small-cell lung cancer. Chest 1985;87:39-43. [Crossref] [PubMed]
- Esscher T, Steinholtz L, Bergh J, et al. Neurone specific enolase: a useful diagnostic serum marker for small cell carcinoma of the lung. Thorax 1985;40:85-90. [Crossref] [PubMed]
- Huang Z, Xu D, Zhang F, et al. Pro-gastrin-releasing peptide and neuron-specific enolase: useful predictors of response to chemotherapy and survival in patients with small cell lung cancer. Clin Transl Oncol 2016;18:1019-25. [Crossref] [PubMed]
- Isgrò MA, Bottoni P, Scatena R. Neuron-Specific Enolase as a Biomarker: Biochemical and Clinical Aspects. Adv Exp Med Biol 2015;867:125-43. [Crossref] [PubMed]
- Girolami I, Lucenteforte E, Eccher A, et al. Evidence-based diagnostic performance of novel biomarkers for the diagnosis of malignant mesothelioma in effusion cytology. Cancer Cytopathol 2022;130:96-109. [Crossref] [PubMed]
- Pisapia P, Pepe F, Iaccarino A, et al. Next Generation Sequencing in Cytopathology: Focus on Non-Small Cell Lung Cancer. Front Med (Lausanne) 2021;8:633923. [Crossref] [PubMed]