
The role of RASA2 in predicting radioresistance in lung cancer through regulation of p53
Highlight box
Key findings
• Ras p21 protein activator (RASA2) could be a potential biomarker for predicting radiosensitivity in lung cancer.
What is known and what is new?
• Mutation or loss of RASA2 promotes RAS activation has been reported in several cancer types, however the role of those mutated RASA2 in therapeutic response remains largely unknown.
• In this study, we found RASA2 limited the efficacy of radiotherapy in lung cancer patients by directly binding to p53 and promoted p53 degradation.
What is the implication, and what should change now?
• RASA2 exhibits potential as a prospective biomarker and genetic target for improving the response to radiotherapy in individuals diagnosed with lung cancer. It is imperative to conduct further preclinical studies to evaluate the efficacy and safety of RASA2 across different cancer types.
Introduction
In advanced non-small cell lung cancer (NSCLC), radiation therapy (RT) with high-energy photons is the first-line option to preventing cancer cell growth in clinical practice and can significantly improve the overall survival and quality of life of patients (1-3). However, for some patients with NSCLC, the response to RT is poor and transient, and the therapy begins to fail several months after its initiation (4). Therefore, in order to provide more personalized treatment, there is an urgent need to better understand radioresistance in patients and discover a new method for stratifying patients based on their different responses (5-7).
Ras p21 protein activator (RASA2), modulates cell proliferation and differentiation (8-10). Previous studies have shown that mutation or loss of RASA2 promotes RAS activation in several cancer types, such as melanoma, prostate cancer, and paraganglioma (8,11,12), indicating its effect on cancer initiation and progression. Nonsynonymous mutations in RASA2 have also been found to be more frequent in patients with lung cancer than in healthy individuals (8); however, the function of RASA2 and its role in therapeutic resistance have not yet been thoroughly examined.
In this study, we found that mutated messenger RNA (mRNA) of RASA2 can be detected in patient tumor tissue, and a higher expression of mutated RASA2 predicted a worse objective response rate (ORR) after 1 month of RT. In terms of the related mechanism, mutated RASA2 was found to directly bind to p53 and promote p53 phosphorylation, with phosphorylated p53 being degraded by the ubiquitin/proteasome pathway. These findings suggest that RASA2 may be an excellent candidate marker for RT prediction and provide insight into the regulatory mechanism of p53. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-160/rc).
Methods
Clinical data
We conducted an observational, retrospective study of 205 patients from January 2016 to June 2018. These patients had performance status (PS) scores below 2, were diagnosed with stage IIIa and IVa lung adenocarcinoma (LUAD), and had never previously received RT. In total, 205 lung cancer tissues were collected by surgical pathologists from Zhongnan Hospital of Wuhan University through computed tomography (CT)-guided needle biopsy. The fresh tumors were kept at 4 ℃ and within 2 hours of collection, the samples were sent to the laboratory. In addition, we stratified patients into a sensitive group [complete response (CR) or partial response (PR)] and an insensitive group [stable disease (SD) or progressive disease (PD)] 1 month after CyberKnife treatment [non-isocentric, 6-megavolt (MV) linear accelerator]. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University (approval No. 2018015K) and informed consent was taken from all the patients.
mRNA expression in patient tissues
Each tumor sample was subjected to RNA extraction following the manufacturer’s instruction (cat no. 217184; Qiagen, Hilden, Germany). After processing, the samples were transferred immediately to storage at −80 ℃. Complement DNA (cDNA) was synthesized from total RNA using the TaqMan Gene Expression Cells-to-CT Kit (cat. no. AM1728; Applied Biosystems, Waltham, MA, USA). Quantitative polymerase chain reaction (qPCR) was performed using TaqMa Fast Advanced Master Mix (cat. no. 4444556; Applied Biosystems, Waltham, MA, USA) as follows: 1 cycle of 2 minutes at 50 ℃ and 20 seconds at 95 ℃ plus 40 cycles of 1 second at 95 ℃ then 20 seconds at 60 ℃. Gene expression was determined by the following formula: mRNA expression level =2−ΔΔCt (13). The sequences of the primers were as follows: RASA2-forward (5'-GCTACCCGATGTCTGGATGA-3'), RASA2-reverse 5'-ATTACAGTGGGGCAGCTCAT-3'); p53-forward (5'-CTCCTCAGCATCTTATCCGAGTG-3'), p53-reverse (5'-GTGGTACAGTCAGAGCCAACC-3'); and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-forward (5'-ACCACAGTCCATGCCATCAC-3'), GAPDH-reverse (5'-TCCACCACCCTGTTGCTGTA-3').
Immunohistochemical (IHC) analysis
Patients’ tumor tissues were fixed in formalin and processed with 30%, 50%, and 70% ethanol, after which they were sent to a histology core facility for embedding and cutting (4 microns per section). The slides were deparaffinized in xylene (2×5 minutes) and rehydrated in 70%, 50%, and 30% ethanol for 5 minutes, respectively. After antigen retrieval, slides were blocked with blocking buffer and incubated with primary antibodies (dilution: 1:300) overnight at 4 ℃. After incubation with secondary anti-rabbit or mouse antibodies (cat. no. BM2004 and BA1038; Boster Bio, Pleasanton, CA, USA) for 1 hour at room temperature, slides were developed with 3,3'-diaminobenzidine (cat no. SK4105; Vector, Newark, CA, USA) for 2 minutes and counterstained with hematoxylin (cat no. H3404; Vector) for 30 seconds, with the reaction being stopped by immersion in water. Slides were then dehydrated (5 minutes each of 95% and 100% ethanol and xylene treatment), covered with Cytoseal 60 (cat no. 23-244257, Fisher Scientific, Hampton, NH, USA) and examined under a light microscope.
Cell culture
HCT116 p53(+/+) (cat. no. C1125) and HCT116 p53(−/−) (cat. no. C1809) were purchased from Whelab (https://whelab.com). NCI-H226 (cat. no. CRL-5826), A549 (cat. no. CRM-CCL-185), HEK293T (cat. no. CRL-1573), NCI-H292 (cat. no. CRL-1848), and HCC827 (cat. no. CRL-2868) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in RPMI 1640 (cat. no. A1049101, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) or Dulbecco’s modified Eagle medium (DMEM; cat. no. 12430054; Gibco) containing 10% fetal bovine serum (cat. no. 10438026; Gibco) and 1% penicillin/streptomycin (cat. no. PYG0016; Boster Bio).
Small interfering RNA (siRNA), short hairpin RNA (shRNA), and plasmids
Human RASA2 siRNA-1 (AUUCAGAGGUUUCAGGGUAA) and RASA2 siRNA-2 (ACUAAAGAGUCCAGUGGUA) were purchased from Invitrogen (Thermo Fisher Scientific), and human RASA2 shRNA (sc-41704-SH) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). The monolayer cells were transfected and screened according to the manufacturer’s instructions.
To introduce the 175H and 273H mutations into the p53 pCB6+ constructs, we performed site-directed mutagenesis by using NEB’s Q5® Site-Directed Mutagenesis Kit (NEB #E0554) according to the manufacturer instructions. The following oligonucleotides were used for 175H: forward primer: AGC GAG GTT GTG AGG CAC TGC CCC CAC CAT GAG CGC TGC CCC CAC CAT GAG CGC TGC T; reverse primer: AGC AGC GCT CAT GGT GGG GGC AGT GCC TCA CAA CCT CCG T. Meanwhile, the following oligonucleotides were used for 273H: forward primer: GGA ACA GCT TTG AGG TGC CATG TTT GTG CCT GTC CTG G; reverse primer: CCA GGA CAG GCA CAA ACA TGC ACC TCA AAG CTG TTC C. Briefly, we first performed exponential amplification to get the PCR product, then PCR product was treated with Kinase, Ligase & DpnI (KLD) reaction for up to 1 hour at room temperature. Five µL of KLD MIX was transformed into 50 µL competent cells (Takara, Kusatsu, Japan; cat. no. 636763), incubated on ice for 30 minutes, then we performed heat shock at 42 ℃ for 30 seconds, expanded the cells by adding 950 µL SOC, gently shake at 37 ℃ for 1 hour. Two hundred and fifty µL of SOC media was spread onto ampicillin agar plate (100 µg/mL), incubated overnight at 37 ℃. The single colony was picked up the next day and performed miniprep using QIAprep Spin Miniprep Kit according to manufacturer instructions (Qiagen; cat. no. 27104). Then the vector was sent to sequence.
For generation cell lines with gain-of-function (GOF)-mutant p53, an empty vector of pCB6+ or constructs expressing p53 175H or 273H (in the PCB6+ vector with the 72R polymorphism) were used and transfected with effectene reagent (Qiagen; cat. no. 301425). Subsequently, cells were selected using G418 (600 µg/mL). The efficacy was confirmed by western blot.
Western blotting analysis and antibodies
Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with blocking buffer [5% slim milk in tris-buffered saline with Tween20 (TBST)] for 1 hour at room temperature and stained with primary antibodies at 4 ℃ overnight. The membrane was then washed in TBST (3× 5 minutes) and incubated with secondary anti-mouse or anti-rabbit antibodies (Santa Cruz Biotechnology) for 1 hour at room temperature. Finally, the membrane was developed by using an enhanced chemiluminescence kit (Invitrogen) and visualized with the Odyssey XF (LI-COR Biosciences, Lincoln, NE, USA). Antibodies used were RASA2 (cat. no. HPA035375; RRID:AB_2674596; Atlas Antibodies, Stockholm, Sweden), p53 [cat. no. 9282; RRID:AB_331476; Cell Signaling Technology (CST), Danvers, MA, USA] , p21 (cat. no. 2947; 2947S, 2947P; RRID:AB_823586; CST), maspin (cat. no. 9117; RRID:AB_2186312; CST), PUMA (cat. no.14570; RRID:AB_2798517; CST), noxa (cat. no. 14766; RRID:AB_2798602; CST), Fas (cat. no. 4233; RRID:AB_2100359; CST), cleaved caspase-3 (cat. no. 9661, NYUIHC-314, 9661S, 9661L; RRID:AB_2341188; CST), cleaved PARP (cat. no. 9541, 9541S, and 9541L; RRID:AB_331426; CST), GAPDH (cat. no. 97166; RRID:AB_2756824; CST), lamin A (cat. no. 13448; RRID:AB_2798221; CST), β-actin (cat. no. 8457, 8457S, 8457L; RRID:AB_10950489; CST), and α-tubulin (cat. no. 2125 and 2125S; RRID:AB_2619646; CST).
Immunoprecipitation and glutathione S transferase (GST) pulldown assay
Cell lysis was carried out using RIPA buffer, and the resulting supernatants underwent initial incubation with protein A/G (MilliporeSigma, Burlington, MA, USA) and the specified antibody overnight at 4 ℃. Subsequently, the precipitates were washed three times with RIPA buffer following centrifugation at 3,000 rpm for 5 minutes. The samples were then subjected to analysis via western blotting. GST-vector or GST-p53 fusion proteins were immobilized on Glutathione SepharoseTM 4B (cat. no. GE17-0756-01; MilliporeSigma) and incubated with lysates derived from HEK293T cells transiently transfected with Flag-tagged RASA2 for 2 hours at 4 ℃. Following five washes, the samples were analyzed through western blotting.
Commercial plasmids
The following plasmids were purchased from Addgene: mutation of 11 prolines to alanines in 64–89 aa region (p53mPRD), pET15b-His-humanp5381–94aa, pET15b-His-human p53, pET15b-His-human p531–320aa, and pET15b-His-human p5394–312aa. The monolayer cells were transfected and screened according to the manufacturer’s instructions.
In vitro ubiquitination assay
NCI-H226 cells were transfected with siRNAs for 48 hours according to the manufacturer’s instructions (Invitrogen). MG132 (10 µM; cat. no. S2619; Selleck Chemicals, Houston, TX, USA) was applied 4 hours before harvesting. The supernatants were coated with anti-Flag beads at 4 ℃ overnight and then analyzed via western blotting.
Assay of cell viability
In the assay of cell viability, 5×103 cells per well were seeded in 96-well plates. These cells were then exposed to the 8-Gy doses of radiation by 6 MV X-rays. After 48 hours, cell toxicity was tested with Cell Counting Kit-8 (cat. no. C0037; Beyotime, Shanghai, China). All experiments were completed as six biological replicates per experiment.
Cell clonogenic survival assays
A549 cells (1×105) were seeded into six-well plates overnight and transfected with the indicated siRNAs. After 48 hours, cells were irradiated with different dosages and then cultured for 2 weeks. After fixation with 10% formalin for 15 minutes room temperature, cells were stained with crystal violet, and a cluster with more than 50 cells was defined as one colony.
Flow cytometry
For cell cycle analysis, 48 hours after transfection with siRNAs, cells were harvested, washed, and stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) for 30 minutes. The cells were then fixed with fixation buffer for anther 30 minutes and examined via flow cytometry (BD FACSCantoTM II, Franklin Lakes, NJ, USA). Data were analyzed by FlowJo v.9 software.
Immunofluorescence staining
Immunofluorescence staining was performed on A549 cells after transfection with the specified siRNAs. These cells were cultured on coverslips and exposed to 4 Gy of irradiation. Subsequently, the cells were fixed with 4% paraformaldehyde (PFA) for 15 minutes at different time points (0 minutes, 30 minutes, 4 hours, and 24 hours) following radiation and permeabilization with 0.2% Triton X-100 for 5 minutes at room temperature. After blocking was performed with 5% bovine serum albumin (BSA) for 1 hour, the samples were subjected to overnight incubation with a γ-H2AX antibody (1:200; cat. no. ab11174; RRID:AB_297813; Abcam, Cambridge, UK), which was followed by a 1-hour incubation with a secondary antibody (1:200; cat no. ab175470; RRID:AB_2783823; Abcam). The cells were stained with then 4',6-diamidino-2-phenylindole (DAPI) for 10 minutes and observed by microscopy.
Animal experiments
Six-week-old male BALB/c nude mice (Beijing HFK Bioscience Co., Ltd., Beijing, China) were kept in a pathogen-free environment. A protocol was prepared before the study without registration. Animal experiments were performed under a project license (No. 143890054R) granted by ethics board of Huazhong University of Science and Technology (HUST), in compliance with HUST institutional guidelines for the care and use of animals. shControl and shRASA2 cells [NCI-H226, HCT116 p53(+/+), HCT116 p53(−/−)] were harvested and resuspended in PBS. Only suspensions consisting of single cells with >90% viability were used for the injections. Subsequently, 5×105 cells in 20 µL of PBS were subcutaneously injected into the lungs of 6-week-old male nude mice. The weight of all mice was weighed two times per week. Bioluminescent imaging (BLI) was conducted for lesions in animal lungs 21 days after tumor implantation, and 24 Gy of irradiation (1.6 Gy/day ×15 days) was initiated following tumor establishment in the lungs of nude mice. The tumors in the lung tissue were then imaged and measured with BLI (IVIS® Spectrum, PerkinElmer, Waltham, MA, USA). For the subcutaneous tumor model, 5×105 cells in 50 µL of PBS were implanted into the right rear flanks of mice. When tumor reached to 40–60 mm3. Irradiation treatment was initiated in the regionally exposed tumor via 40 Gy of X-rays (10 Gy/day × 4 days). Each animal was tracked individually for tumor growth with external caliper measurements, and the approximate tumor volume was calculated using the following formula: length × width2 × 0.5. Each in vivo experiment was repeated three times, and groups of five mice per experiment were used for experimental tumor assays to ensure adequate power in detecting biological differences. Tumor burden did not exceed the recommended dimensions (largest diameter <1.8 cm), and animals were anesthetized with isoflurane and killed via carbon dioxide.
Bioinformatic analysis
To compare the expression of RASA2 in normal lung tissue with that in tumors, we analyzed the lung cancer database from The Cancer Genome Atlas (TCGA) and extracted RNA sequencing (RNA-seq) data from level 3 high-throughput sequencing (HTSeq)-fragments per kilobase of exon model per million mapped reads (FPKM), which were then transformed into log2 format. Data were visualized with the “ggplot2” package in Rstuido (version 3.6.3, Posit PBC, Boston, MA, USA).
To assess RASA2 performance in predicting cancer, we first extracted the corresponding normal tissue data from the Genotype-Tissue Expression (GTEx) and cancer tissue (LUAD) from TCGA. The University of California Santa Cruz (UCSC) Xena browser (https://xenabrowser.net/datapages/) was used to transform RNA-seq data of TCGA and GTEx into transcripts per million reads (TPM) format according to the Toil pipeline (14). The RNA-seq data in TPM was then transformed into log2 format for comparison between samples. Finally, we used the “pROC” (version 1.17.0.1) R package to analyze the data and the “ggplot2” (version 3.3.3) R package for visualization.
Statistical analysis
Results are expressed as the mean ± standard deviation (SD). Receiver operating characteristic (ROC) curves were generated to evaluate sensitivity, specificity, and the corresponding area under the curve (AUC) and the 95% confidence intervals (CIs). The optimal cutoff value for prediction was determined by maximizing the sum of sensitivity and specificity while minimizing the overall error [calculated as the square root of the sum of (1 − sensitivity)2 + (1 − specificity)2]. All analyses were performed using GraphPad Prism 8.0 (GraphPad Software). The significance threshold was set as *, P<0.05; **, P< 0.01; ***, P<0.001.
Results
RASA2 predicted a poor RT response in patients with lung cancer
Since inactivating RASA2 mutations have been reported in some types of cancer (melanoma and multiple myeloma) (8), we first sought to investigate RASA2 expression from available public databases. Bulk RNA-seq data of lung cancer [LUAD/lung squamous cell carcinoma (LUSC)] from the GTEx and TCGA databases showed that compared to normal lung tissue, lung cancer tissues had significantly increased RASA2 expression (Figure 1A, Table S1). Notably, the expression of RASA2 had good performance in predicting malignancy (normal versus tumor) with a high AUC (0.820; 95% CI: 0.791–0.848) (Figure 1B). Therefore, to examine the role of RASA2 in RT for patients, we initiated an open, observational, retrospective study of 205 patients from January 2016 to June 2018. The criteria for enrolment were patients with a PS score below 2 who were diagnosed with stage IIIa and IVa LUAD and had never previously received RT. The features of all patients are displayed in Table S2. Specimens from patients with lung cancer were acquired via CT-guided percutaneous needle biopsy (Figure 1C). IHC staining for RASA2 in human lung cancer tissue showed that RASA2 was located mainly in the cytoplasm (Figure 1D), which is consistent with the data from public databases (Figure 1E).
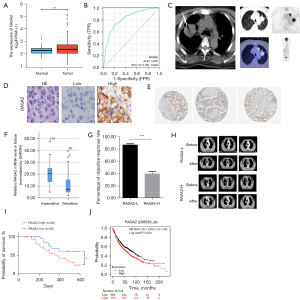
We then utilized primers as described previously (15,16) to clarify the relationship between RASA2 and RT response, and we stratified patients into a sensitive group or an insensitive group. As shown in Figure 1F, patients in the sensitive group had a lower mRNA expression of RASA2 than did those in the insensitive group, which indicated that RASA2 expression may be negatively correlated with the RT response in lung cancer. Additionally, patients with a high expression of RASA2 had a much lower ORR than did those with a low expression of RASA2 (Figure 1G), as evidenced by the larger tumors after irradiation (Figure 1H) and overall survival (Figure 1I). The Kaplan-Meier plot from the publicly available data on patients with lung cancer also showed that patients with high RASA2 expression had shorter overall survival (Figure 1J). Altogether, these results suggest that RASA2 is related to a poor RT response in patients.
Knockdown RASA2 enhanced RT by inducing DNA damage and apoptosis
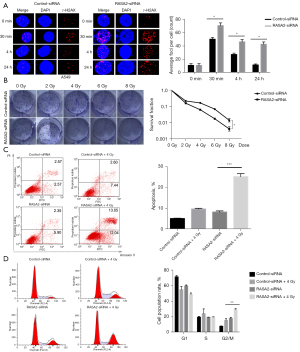
We next sought to determine the role of RASA2 in mediating radioresistance in vitro. To evaluate irradiation-induced DNA damage, we performed a γ-H2AX foci assay and found that knocking out RASA2 robustly increased the number of γ-H2AX foci (Figure 2A). In addition, colony formation was substantially decreased in the RASA2-depleted human lung cancer cell line A549, indicating that knocking down RASA2 promotes the radiosensitivity of cancer cells (Figure 2B). Importantly, this phenotype is mediated by increased apoptosis, as the proportions of apoptotic and G2/M phase cells were significantly increased in RASA2-knockdown cells (Figure 2C,2D). Taken together, these findings suggested that knocking down RASA2 promotes radiosensitivity by inducing apoptosis and DNA damage in lung cancer cells.
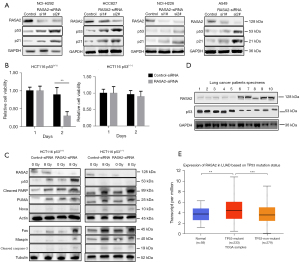
Since p53 plays an important role in processes related to DNA damage and cell death, such as apoptosis, we wondered whether the effects of RASA2 on radioresistance are mediated by p53. We first evaluated p53 expression in wild-type lung cancer cell lines (NCI-H292, HCC827, NCI-H226, and A549). Interestingly, the expression of p53 and its target gene, p21, both increased after knocking down RASA2 (Figure 3A). Next, we knocked down RASA2 in the HCT116 p53(+/+) and HCT116 p53(−/−) cell lines using siRNAs. As expected, irradiation significantly inhibited cell proliferation in the p53-proficient HCT116 cell line but not in the p53-deficient cell line (Figure 3B). In addition, 72 hours after 8-Gy irradiation, we tested p53-related proteins in RASA2-knockdown HCT116 p53(+/+) and HCT116 p53(−/−) cells. In the HCT116 p53(+/+) cell line, knocking down RASA2 increased the expression of p53 and apoptosis-related proteins (maspin, PUMA, noxa, Fas, cleaved caspase-3, and PARP), while there were no differences in the HCT116 p53(−/−) cell line (Figure 3C), indicating that RASA2 may cause radioresistance by interrupting p53. Next, we aimed to determine whether RASA2 can affect the cell lines with the GOF features due to the p53 mutations, p53 175H or p53 273H, which are frequently found in human cancers (17). In our investigation into the potential impact of RASA2 on cell lines featuring GOF characteristics resulting from p53 mutations, specifically p53 175H or p53 273H, commonly observed in various human cancers (17). To simulate these conditions, we introduced the aforementioned p53 mutants into our HCT 116 p53(−/−) cancer cell line (Figure S1A). It’s worth noting that the HCT 116 p53(−/−) cell line lacks endogenous p53 expression, a crucial factor in ensuring that any discernible effects attributed to the mutant p53 variants would not be influenced by dominant-negative inhibition of wild-type p53. Contrary to our initial expectations, the inhibition of RASA2 did not yield the anticipated restoration of radiosensitivity in our experimental setup, as evidenced by the assessment of colony formation (Figure S1B). Additionally, RASA2 inhibition did not result in a significant increase in apoptosis, as shown in Figure S1C. These unexpected findings suggest a nuanced relationship between RASA2 and p53 functionality. Notably, it appears that RASA2 may predominantly exert its influence in a negative manner when the function of p53 is intact. Moreover, upon further investigation of tumor tissue from patients, we found that RASA2 was negatively correlated with p53 in samples of patients before they received irradiation (Figure 3D). Interestingly, analysis of available public databases revealed that patients with p53 mutations [missense mutations (75%), frameshift insertion and deletion (9%), nonsense mutation (7%), silent mutation (5%), and rare changes] had much higher RASA2 gene expression levels than did patients with the p53 wild type (Figure 3E). However, most of the mutations were missense, and thus for special mutations other than GOF, the role of RASA2 in regulating p53 requires further scrutiny.
RASA2 regulated p53 stability
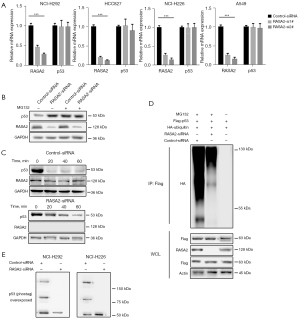
RASA2 deficiency did not affect transcriptional activity of p53 (Figure 4A) but upregulated p53 expression in the cytoplasm, which allowed us to ascertain whether RASA2 affects p53 protein stability at the posttranslational level. p53 is degraded by the proteasome, and we also assessed whether RASA2 affects p53 through the ubiquitin/proteasome pathway. Interestingly, after treatment with the proteasome inhibitor MG132, protein levels of p53 remained stable in RASA2-knockdown cells (Figure 4B), suggesting that loss of RASA2 prevented p53 degradation through the ubiquitin/proteasome signaling pathway. In addition, we also observed a longer half-life of endogenous p53 protein in RASA2-knockdown cells (Figure 4C). Importantly, p53 polyubiquitination was decreased when RASA2 was knocked down (Figure 4D), suggesting that RASA2 was dependent on ubiquitination to degrade p53.
RASA2 binding with p53 via the proline-rich domain
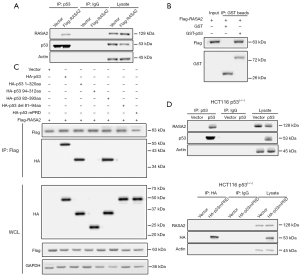
We next assessed how RASA2 regulated p53 stability. Posttranslational modifications (PTMs), which mainly include phosphorylation, acetylation, and ubiquitination, are intricate patterns that regulate p53. Our previous study showed that phosphorylation of p53 at the serine 15 site by ataxia telangiectasia mutated (ATM) kinase leads to p53 stabilization during DNA damage (18); therefore, we first assessed whether a similar pattern is involved in changes in RASA2 expression. Interestingly, we also observed reduced p53 phosphorylation in RASA2-knockdown cell lines (Figure 4E), and this change was mainly observed in the cytoplasm (Figure S2A,S2B). We next examined whether RASA2 could directly interact with p53. Unexpectedly, RASA2 was coimmunoprecipitated with p53 (Figure 5A); we performed a GST pulldown assay and discovered that p53 was immunoprecipitated with RASA2 (Figure 5B), indicating that RASA2 can directly interact with p53 in vitro. p53 is a protein with a multifunctional domain, and different domains play different roles in phosphorylation and other PTMs. Common mutants of human p53 were evaluated for binding to RASA2; these included p531–320aa, p5382–393aa, p5394–312aa, p53del81–94aa, and p53mPRD (conversion of 11Ps to 11As in the PRD). RASA2 bound to p531–320aa and p5382–393aa but failed to bind to p5394–312aa, indicating that RASA2 may bind to the p5382–94aa region. In addition, p53 with deletion of the PRD (p53del81–94aa) or with mutation of the PRD (p53mPRD) could not bind to RASA2 at all. This indicated that RASA2 directly binds to the PRD in p53 (Figure 5C), and this was confirmed in HCT116 p53(−/−) cells; as expected, the PRD-mutant p53 protein failed to bind to RASA2 (Figure 5D), confirming that RASA2 can directly bind with p53 in the PRD.
Downregulation of RASA2 enhanced the RT response within tumors
Our in vitro results suggested that RASA2 promotes radioresistance by regulating p53 protein modification and degradation. To recapture the mechanism in vivo, we generated an orthotopic tumor model that could better represent the real tumor microenvironment than could a subcutaneous tumors model. BLI at 14 and 30 days after irradiation indicated that knocking down RASA2 reduced tumor growth (Figure 6A,6B). To demonstrate that the efficacy of knocking down RASA2 is mediated by p53, we performed mouse experiments by knocking down RASA2 in both HCT116 p53(+/+) and HCT116 p53(−/−) cells. After 40 Gy of radiation treatment, mice bearing RASA2-deficient cancer cells had much smaller tumors than the control mice in the HCT116 p53(+/+) group. Nevertheless, no significant changes were observed between the two groups for HCT116 p53(−/−) (Figure 6C). In line with this, apoptosis-related protein levels were increased in RASA2-knockdown mice as compared with RASA2 control mice in the HCT116 p53(+/+) group, while there was no related difference in the HCT116 p53(+/+) group (Figure 6D). Together, these data suggest that knocking down RASA2 enhances the irradiation response in vivo.

Discussion
In the lung cancer field, a large proportion of patients eventually develop resistance to RT, which leads to cancer progression (19). In this study, we found that RASA2 acts as an independent predictor of the RT response in patients with lung cancer. Mechanistically, RASA2 destabilizes p53 by directing binding with p53, which further accelerates p53 degradation and blocks the p53-related apoptosis pathway.
RASA2 plays an important role in suppressing RAS activity. However, in recent years, loss-of-function (LOF) mutations of RASA2 have been implicated in some types of cancers, especially multiple myeloma and melanoma (8). The general understating is that mutated RASA2 cannot negatively regulate the MAPK signaling pathway, which leads to uncontrolled tumor growth. There are some new views that Ras p21 coordinates with guanine nucleotide-binding regulatory proteins, which subsequently regulates cellular adenyl cyclase and the process of intracellular phosphorylation, which may undergo amplification in both the processes of ontogenesis and oncogenesis (11,20,21).
In this study, we used public databases to investigate RASA2 expression. To our surprise, in lung cancer, RASA2 expression was significantly increased compared with that in normal tissue. Moreover, RASA2 showed good performance in distinguishing normal versus tumor tissues. Based on these findings, we initiated a retrospective clinical study by collecting patients’ tissue samples before and after RT. For the first time, we observed that RASA2 mRNA levels were negatively correlated with RT response, and patients with high RASA2 levels were more resistant to RT. The principal mechanism of RT resistance is closely related to DNA damage, and we thus assessed whether knocking down RASA2 had any effect on DNA damage in cells. As expected, knocking down RASA2 significantly induced DNA damage, as indicated by γ-H2AX and the cell apoptosis pathway (maspin, PUMA, noxa, and Fas). p53 is a master tumor-suppressor protein in the cell cycle that acquires LOF mutations during cancer progression and treatment resistance (22,23). Previously, we showed that human ssDNA binding protein SSB1 (hSSB1) interacts with p53 through regulating transcription and stability (24). Therefore, we sought to determine whether the effect of RASA2 is mediated by p53. We first found that knocking down RASA2 could stabilize p53 protein expression through PTM. Importantly, RASA2 can phosphorylate p53 by binding to p53 at the PRD, which can lead to destabilization of p53 and inhibit p53 transcription. Finally, we verified these findings in an orthotopic lung cancer animal model. After irradiation, mice with RASA2-knockdown cells had significantly smaller tumors than did the mice in the control group. This finding is not limited to a single mouse model. Interestingly, in the HCT116 p53(+/+) group, RASA2-knockdown mice had smaller tumors than did the control group after irradiation. However, there was no difference in the tumor control between the RASA2-knockdown and control mice in the HCT116 p53(−/−) group, indicating that the effect of RASA2 is mediated by p53 (Figure 6E).
Interestingly, a recent study showed that RASA2 ablation in T cells boosted antigen sensitivity and long-term function, which points to the dual role of RASA2 in tumor and immune cells (9). Thus, targeting RASA2 may have a twofold effect.
Initially, it’s important to highlight that RASA2 frequently co-occurs with mutations in other tumor suppressors, such as neurofibromatosis type 1 (NF1). For example, in melanoma, loss of RASA2 and NF1 have been found to have complementary pro-tumorigenic functions (12). This co-mutation suggests that focusing solely on RASA2 might not be effective, due to the interconnected nature of these pathways, potentially limiting the broader applicability of our findings. Another limitation arises from the methodology used to study the RASA2 mutation profile. By relying on primer sequences from pre-existing research (16), our approach may not capture the complete mutation landscape of RASA2. This reliance potentially limits the accuracy and comprehensiveness of our findings, leaving the true mutation frequency and spectrum of RASA2 only partially understood. Advancements in sequencing technologies and more bespoke primer design could alleviate this issue, offering deeper insights into RASA2’s role in cancer. Lastly, the exclusion of the immune system’s role from our study represents a significant oversight, given the growing recognition of immuno-oncology in cancer therapy. Specifically, the potential synergies between immune checkpoint inhibitors and RASA2 mutations were not explored. This area of research could unveil novel therapeutic strategies by modulating immune response to fight cancer more effectively.
Conclusions
Our research underscores the frequent modifications in RASA2, offering an alternative explanation for the disruption of apoptosis in cancer cells. RASA2 shows promise as both a potential biomarker and a genetic target to enhance the response to RT in patients with lung cancer. Based on TCGA datasets of LUAD and LUSC, it was observed that there are no significant differences in the expression levels of RASA2 between male and female lung cancer patients. This finding is particularly intriguing as it suggests that the expression of RASA2 in lung cancer is likely not influenced by hormonal differences between genders, such as those attributed to estrogen and testosterone. Typically, hormones play significant roles in the progression and development of lung cancer, and their lack of influence on RASA2 expression warrants further investigation. In addition, we found no statistical significance in RASA2 expression levels across the various stages of lung cancer (data not shown). This uniformity in expression levels from early to advanced stages of the disease suggests that RASA2’s expression is consistent throughout the progression of lung cancer. This consistency underscores the potential importance of early detection of RASA2 expression in patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-160/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-160/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-160/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-160/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University (approval No. 2018015K) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 143890054R) granted by ethics board of Huazhong University of Science and Technology (HUST), in compliance with HUST institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vinod SK, Hau E. Radiotherapy treatment for lung cancer: Current status and future directions. Respirology 2020;25:61-71. [Crossref] [PubMed]
- Li H, Zhao Y, Ma T, et al. Radiotherapy for extensive-stage small-cell lung cancer in the immunotherapy era. Front Immunol 2023;14:1132482. [Crossref] [PubMed]
- Wu Y, Song Y, Wang R, et al. Molecular mechanisms of tumor resistance to radiotherapy. Mol Cancer 2023;22:96. [Crossref] [PubMed]
- Galeaz C, Totis C, Bisio A. Radiation Resistance: A Matter of Transcription Factors. Front Oncol 2021;11:662840. [Crossref] [PubMed]
- Pennell NA, Arcila ME, Gandara DR, et al. Biomarker Testing for Patients With Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am Soc Clin Oncol Educ Book 2019;39:531-42. [Crossref] [PubMed]
- Binkley MS, Jeon YJ, Nesselbush M, et al. KEAP1/NFE2L2 Mutations Predict Lung Cancer Radiation Resistance That Can Be Targeted by Glutaminase Inhibition. Cancer Discov 2020;10:1826-41. [Crossref] [PubMed]
- Zhou E, Li Y, Wu F, et al. Circulating extracellular vesicles are effective biomarkers for predicting response to cancer therapy. EBioMedicine 2021;67:103365. [Crossref] [PubMed]
- Arafeh R, Qutob N, Emmanuel R, et al. Recurrent inactivating RASA2 mutations in melanoma. Nat Genet 2015;47:1408-10. [Crossref] [PubMed]
- Carnevale J, Shifrut E, Kale N, et al. RASA2 ablation in T cells boosts antigen sensitivity and long-term function. Nature 2022;609:174-82. [Crossref] [PubMed]
- Johansen KH, Golec DP, Okkenhaug K, et al. Mind the GAP: RASA2 and RASA3 GTPase-activating proteins as gatekeepers of T cell activation and adhesion. Trends Immunol 2023;44:917-31. [Crossref] [PubMed]
- Hamdy S, Aprikian A, Begin L, et al. Ras p21 overexpression is a late event in prostate-cancer. Int J Oncol 1994;4:627-31. [Crossref] [PubMed]
- Arafeh R, Di Pizio A, Elkahloun AG, et al. RASA2 and NF1; two-negative regulators of Ras with complementary functions in melanoma. Oncogene 2019;38:2432-4. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 2017;35:314-6. [Crossref] [PubMed]
- Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell 2012;150:251-63. [Crossref] [PubMed]
- Jiang M, Shi X, Zhu H, et al. Two GEO MicroRNA Expression Profile Based High-Throughput Screen to Identify MicroRNA-31-3p Regulating Growth of Medullary Thyroid Carcinoma Cell by Targeting RASA2. Med Sci Monit 2019;25:5170-80. [Crossref] [PubMed]
- Schulz-Heddergott R, Stark N, Edmunds SJ, et al. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018;34:298-314.e7. [Crossref] [PubMed]
- Xie J, Li Y, Jiang K, et al. CDK16 Phosphorylates and Degrades p53 to Promote Radioresistance and Predicts Prognosis in Lung Cancer. Theranostics 2018;8:650-62. [Crossref] [PubMed]
- Césaire M, Montanari J, Curcio H, et al. Radioresistance of Non-Small Cell Lung Cancers and Therapeutic Perspectives. Cancers (Basel) 2022;14:2829. [Crossref] [PubMed]
- Rajalingam K, Schreck R, Rapp UR, et al. Ras oncogenes and their downstream targets. Biochim Biophys Acta 2007;1773:1177-95. [Crossref] [PubMed]
- Davidson B, Agulansky L, Goldberg I, et al. Immunohistochemical analysis of rasGTPase activating protein (rasGAP) in prostate cancer. Pathol Res Pract 1998;194:399-404. [Crossref] [PubMed]
- Stewart-Ornstein J, Iwamoto Y, Miller MA, et al. p53 dynamics vary between tissues and are linked with radiation sensitivity. Nat Commun 2021;12:898. [Crossref] [PubMed]
- Kong X, Yu D, Wang Z, et al. Relationship between p53 status and the bioeffect of ionizing radiation. Oncol Lett 2021;22:661. [Crossref] [PubMed]
- Xu S, Wu Y, Chen Q, et al. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res 2013;23:423-35. [Crossref] [PubMed]


