
Regulatory role of the miR-142-3p/CDC25C axis in modulating autophagy in non-small cell lung cancer
Highlight box
Key findings
• Our study identified cell division cycle 25C (CDC25C) as an important prognostic marker in non-small cell lung cancer (NSCLC), emphasizing the role of the miR-142-3p/CDC25C axis in the regulation of fundamental cellular processes such as proliferation, apoptosis and autophagy. This emphasizes the discovery of the miR-142-3p/CDC25C axis and its wide-ranging impact in NSCLC.
What is known and what is new?
• The progression of NSCLC is influenced by complex genetic interactions, and understanding these can inform prognosis and treatment.
• This manuscript establishes the miR-142-3p/CDC25C axis as a targetable mechanism for therapeutic intervention and exposes the previously unknown effect of CDC25C on the advancement of NSCLC.
What is the implication, and what should change now?
• The findings suggested that targeting the miR-142-3p/CDC25C axis may be a promising therapeutic strategy for NSCLC, and further studies are needed to explore the therapeutic potential of this regulatory axis in NSCLC.
Introduction
Malignancy that develops in the bronchial mucosa or glandular tissue of the lungs is known as lung cancer, or pulmonary carcinoma (1). Non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) constitute the two primary forms of lung cancer (2). NSCLC accounts for 80–85% of all cases of lung cancer, establishing it as a predominant subtype within the spectrum of lung carcinomas (3). NSCLC is primarily caused by exposure to risk factors such as smoking, secondhand smoke, occupational carcinogens, and environmental pollutants, which can lead to the development of genetic mutations and cellular abnormalities in the lung (4). Lung cancer exhibits astonishingly high incidence and mortality rates, making it a major global public health concern. Since the early symptoms are so inconspicuous, many individuals are diagnosed at an advanced stage of illness, which reduces the chance of an effective course of treatment (5). Molecular translational research has made remarkable progress in the field of NSCLC, with impressive success in targeted therapy in particular. A study had emphasized the comprehensive exploration of tumor target antigens and related drugs, providing important theoretical support for NSCLC treatment. In addition, summaries of clinical trial studies conducted on NSCLC patients have provided strong support to drive further development of treatment strategies (6). Nonetheless, the current situation was that some patients can obtain a better prognosis through early diagnosis and personalized treatment, the prognosis of most patients differs due to factors such as stage, subtype, and molecular characteristics (7). Therefore, targeted therapy research for lung cancer patients is urgently needed to better understand the characteristics of different patient groups and to improve the therapeutic efficacy, which is an urgent need for current medical research (8).
Cell division cycle 25C (CDC25C) serves as a pivotal regulator of cyclin-dependent kinases (CDKs) within the cell cycle, primarily overseeing cell cycle progression during the transition from G2 to M phase before mitosis initiation (9). It appears that CDC25C might be a valuable target for cancer treatment, as the expression of this gene has been consistently elevated in a number of cancers, including stomach (10), lung (11), and bladder cancer (12). CDC25C also holds substantial significance within NSCLC research due to its multifaceted applications. Previous study had shown that the CDC25C-related DNA damage repair (DDR) gene prognostic model was established through DDR and found that CDC25C may be a potential therapeutic marker for NSCLC (13). Furthermore, heightened CDC25C expression in NSCLC patients has been correlated with unfavorable clinical outcomes following targeted drug interventions, reinforcing the pivotal role of CDC25C (14). Recent study had substantiated the inhibitory effects of Cyclovirobuxine D (CVB‑D) on the growth and advancement of NSCLC cells. This inhibition was attributed to the modulation of the KIF11‑CDK1‑CDC25CCyclinB1 G2/M phase transition regulatory network and the suppression of the NF-κB/JNK signaling pathway (15). This, in turn, offers a promising avenue for targeted lung cancer therapies. Nevertheless, the precise biological functions and mechanisms through which CDC25C operates in NSCLC remain elusive. In light of its crucial function in controlling the cell cycle and its possible use as a therapeutic target, further research must be conducted to clarify its roles and molecular complexities in lung cancer.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that play key roles in human cancers (16). Studies had shown that aberrant expression of miRNAs may be a potential target for cancer therapy covering a wide range of cancer types, such as breast cancer (17), prostate cancer (18), and colorectal cancer (19). In particular, miR-142-3p is considered a tumor suppressor that plays an important role in a variety of cancers. For example, miR-142-3p has been shown to inhibit the proliferation and invasion of pancreatic cancer cells (20). Furthermore, miR-142-3p has been shown to promote cell proliferation in human nasopharyngeal carcinoma by inhibiting SOCS68 (21). However, although miR-142-3p has been shown to play an important role in some cancer types, its potential function in NSCLC requires further in-depth studies.
Given the complex molecular mechanism revealed by comprehensive NSCLC analyses, the goal of this research was to identify potential targets that drive tumor growth and prognosis. To identify potential prognostic targets, we examined differentially expressed genes (DEGs) in the GSE68571 and The Cancer Genome Atlas (TCGA)-NSCLC datasets using bioinformatics techniques. The results confirmed the existence of CDC25C as a stand-alone prognostic factor. Subsequently, the dual roles of CDC25C in regulating NSCLC cell phenotypes were examined through in vitro assays. Besides, the regulatory influence of miR-142-3p on these processes was investigated. Our results shed light on the complex biological pathways involved in NSCLC that are influenced by the miR-142-3p and CDC25C regulatory networks. We present this article in accordance with the TRIPOD and MDAR reporting checklists (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-82/rc).
Methods
Comparative analysis of differential gene expression in NSCLC using TCGA and Gene Expression Omnibus (GEO) datasets
We retrieved 1,017 NSCLC samples and 108 normal samples from the TCGA database (https://tcga-data.nci.nih.gov/tcga/). For further confirmation, we selected the GSE68571 dataset in the GEO (https://www.ncbi.nlm.nih.gov/gds/) database, which included 86 samples from lung cancer individuals and ten samples from non-lung cancer individuals. We performed differential expression analysis on this set of data using the “Limma” package of R program (The R Foundation for Statistical Computing, Vienna, Austria). To ensure the reliability of our findings, we went through both datasets with the same set of filtering standards. Genes were filtered based on fold change (FC) standards, specifically, DEGs with FC >2 were considered upregulated, and those with FC <0.5 were categorized as downregulated, and a significance criterion of P<0.05 was used to determine statistical significance. Venn analysis was used to reveal which genes among the up- and down-regulated DEGs in the two databases were overlapping. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Protein-protein interaction (PPI) analysis and functional enrichment
To create a PPI network for proteins encoded by DEGs, we used the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/). We employed the Molecular Complex Detection (MCODE) method, a plugin that can be accessed through Cytoscape software (https://cytoscape.org/), to find highly linked submodules inside the PPI network. The “ClusterProfiler” package performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) term enrichment analyses for genes in selected submodules. Molecular function (MF), cellular component (CC), and biological process (BP) are the three hierarchical categories into which GO categorization splits gene and protein functions. KEGG pathways are typically employed to elucidate the roles of genes in various metabolic pathways and BPs. Enrichments with P values <0.05 were deemed statistically significant.
Least absolute shrinkage and selection operator (LASSO) analysis
To generate a prognostic model and identify potential candidate genes, we employed the “glmnet” package within the R software. We performed 10-fold cross-validation on LASSO regression analysis to determine the significant gene coefficients. For each NSCLC patient, this formula was used to determine a risk score: risk score = −0.1693 × EZH2 + 0.112 × CDC25C + 0.0415 × TYMS − 0.0756 × RFC4 + 0.0483 × KIF14 + 0.1469 × PLK1 + 0.049 × CCNE1. NSCLC patients were stratified according to median risk score. To assess the predictive performance of the identified significant genes in NSCLC, we utilized the “survival” package in R to generate Kaplan-Meier (KM) survival curves, allowing for a comparison of differences in overall survival (OS) probabilities. The significance of the differences in survival was assessed using the log-rank test. Area under the curve (AUC) values were calculated through receiver operating characteristic (ROC) curves created using the “timeROC” R package to further analyze the prognostic characteristics of candidate genes.
Comprehensive prognostic assessment and differential expression analysis in NSCLC
Using the “forestplot” package, univariate and multivariate Cox regression analyses were carried out on prognostic genes and clinical variables to evaluate the prognostic importance of various parameters. The “rms” package generates nomograms to predict survival probabilities at 1, 3, and 5 years for important clinical variables. A visual depiction of the predicted odds of surviving over time was offered by these nomograms. The link between anticipated and actual survival results was shown using calibration curves, which were created to evaluate the accuracy and effectiveness of nomograms. The expression levels of a key prognostic gene in the nomogram were then determined in TCGA-NSCLC samples and the GSE68571 dataset, respectively.
Cell culture
The Shanghai Cell Institute in China provided the NSCLC cell lines: Calu-3 [American Type Culture Collection (ATCC; Manassas, VA, USA), HTB-55], A549 (ATCC, CRM-CCL-185), H1975 (ATCC, MD, CRL-5908), H2228 (ATCC, CRL-5935) as well as the human embryonic lung fibroblast cell line MRC-5 (ATCC, Cat #CCL-171). Cell maintenance followed established protocols detailed by Lachowicz et al. [2019] (22). Cells were treated with different doses of 3-methyladenine (3-MA) (0, 5, 10, 20 µM) for 24 hours to study the relationship between the hub gene and autophagy. In addition to the above cell lines, we also performed dual-luciferase assays using 293T cells derived from the ATCC.
Cell transfection
Logarithmic growth stage NSCLC cells were collected, diluted, and seeded into a density of 2×106 cells on a six-well plate. Upon reaching 50–60% cell confluence, transfection was performed. The miR-142-3p inhibitor, miR-142-3p mimics, si-CDC25C-1, si-CDC25C-2, and their respective negative controls were designed and provided by GenePharma (Shanghai, China). Transfecting the cells with the plasmids indicated above was accomplished using the Lipofectamine 2000 transfection kit (Invitrogen, Carlsbad, CA, USA). The initial medium was replaced with a complete media after a 6-hour culture. The cells were then collected 48 hours following transfection.
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used to extract total RNA from NSCLC cells in accordance with the manufacturer’s instructions. Subsequently, complementary DNA (cDNA) synthesis was carried out using PrimeScript RT Master Mix (Takara, Dalian, China) according to the manufacturer’s protocol. For qRT-PCR, SYBR Green PCR Master Mix from (Takara, China) was utilized, and the reactions were conducted on an ABI 7900HT instrument [Applied Biosystems, Foster City, CA, USA]. For messenger RNA (mRNA) analysis, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) acted as the internal control. Meanwhile, the miR-142-3p expression study used U6 as the internal reference. The primer sequences for CDC25C were as follows: forward 5'-GTGTTCCTCTGTGAATTCT-3', reverse 5'-CGTGTTCCATTGCGAGTTCTC-3'. The primer sequences for miR-142-3p were as follows: forward 5'-GTCGTATCCAGTGCAGGG-3', reverse 5'-CGACGTGTAGTGTTTCCCTA-3'. GAPDH forward primer: 5'-CAAGCTCATTTCCTGGTATGAC-3' and reverse primer: 5'-CAGTGAGGGTCTCTCTCTTCCT-3'. U6 forward primer: 5'-CGCTTCGGCAGCACATATAC-3', and reverse primer: 5'-CAGGGGCCATGCTAATCTT-3'. The 2−ΔΔCt technique was utilized to examine the expression of miR-142-3p and CDC25C.
Western blotting (WB) assay
In order to create protein lysates from MRC-5, A549, Calu-3, H1975, and H2228, protease and phosphatase inhibitors were added to radioimmunoprecipitation assay (RIPA) lysis solution (Thermo Fisher Scientific). The bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) was used to measure the amounts of proteins. After sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for electrophoresis, polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA, USA) were transferred. Immunoblotting was carried out using primary antibodies against CDC25C, Bax, Bcl-2, LC3 I, LC3 II, p62, cyclin B, CDK1-Y15, CDK1, c-PARP, and GAPDH (all at 1:1,000; Abcam, Cambridge, MA, USA). Post-washing thrice with tris-buffered saline with Tween 20 (TBST), membranes were processed for band visualization using an enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific). Using the ChemiDoc system from Bio-Rad (Hercules, CA, USA), the intensities of the protein bands were recorded and subsequently analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cell counting kit-8 (CCK-8) assay
The CCK-8 kit (CCK-8, Hanbio, Shanghai, China) was used to assess cell viability according to the manufacturer’s instructions. NSCLC cells were cultured in 96-well plates (5×103 cells/well) with 10% fetal bovine serum (FBS) added to the medium in six replicates per sample. Ten µL of CCK-8 reagent was added to each well. Cell viability was determined by measuring absorbance at 450 nm every 24 hours using a Thermo Fisher microplate reader. Data represent the results of independent experiments performed in triplicate.
Transwell assays
After 24 hours of transfection, cells were taken out and reconstituted at a 5×104 cell/well concentration. These cells were then placed into the upper chamber of a Transwell insert (six-well format). Complete medium was introduced to the lower chamber to act as a chemoattractant. The cells that remained in the upper chamber and did not migrate or invade were carefully removed using a cotton swab after a 48-hour incubation period at 37 ℃. The nuclei of the cells on the underside of the membrane were visible by staining with a crystal violet cell colony staining kit after the cells had been fixed with 4% paraformaldehyde. Using a fluorescent microscope (Nikon, Tokyo, Japan), the cells that had migrated or invaded were then observed and counted. Following that, photographs were taken for documentation.
Flow cytometry assay
After collection, the cells were washed in phosphate-buffered saline (PBS) and left to fix in 70% ethanol for the night at −20 ℃. An annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit was used to stain cells for determination of apoptosis. The cells were treated with RNase A and then stained for 30 minutes at 37 ℃ in the dark using 50 g/mL PI. The PI staining was then utilized to assess the distribution of the cell cycle. The FACSCalibur [Becton, Dickinson, and Co. (BD) Biosciences, Franklin Lakes, NJ, USA] was used to gather the data, which were then processed with FlowJo software (BD Biosciences).
Dual-luciferase reporter assay
Based on bioinformatics predictions, the CDC25C 3'-untranslated region (3'-UTR)-predicted binding sequences for miR-142-3p were amplified and inserted into pmirGLO vectors (Promega, Madison, WI, USA). Using Lipofectamine® 3000, the miR-142-3p mimic and both CDC25C wild-type (WT) and mutant-type (MUT) luciferase reporter vectors were co-transfected. Luciferase activity was assessed 48 hours later using a dual-luciferase system (Promega).
Colony formation assay
Counts of 2,000 cells per group were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% FBS in six-well plates for 10 days. After washing the cells with PBS, the cells were stained using a crystal violet cell colony staining kit (Genemed, Shanghai, China). Under a microscope, colonies were counted and images were captured using a colony formation assay.
Statistical analysis
Data were analyzed using SPSS 18.0 (IBM Corp., Chicago, IL, USA), showing mean ± standard deviation (SD) for triplicate experiments. Student’s t-test was used to assess group difference, and a P value of less than 0.05 was considered statistically significant.
Results
PPI and enrichment analysis of overlapping genes in NSCLC
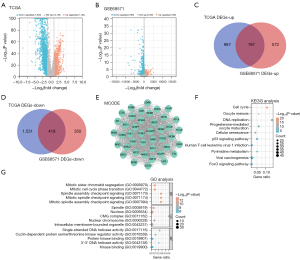
Differential expression analysis was performed on genes from both the TCGA database and the GSE68571 dataset. In the TCGA dataset, we identified 1,184 upregulated DEGs and 1,950 downregulated DEGs (Figure 1A) using the limma R software package. Similarly, in the GSE68571 dataset, we detected 769 upregulated DEGs and 769 downregulated DEGs (Figure 1B). We further employed the “VennDiagram” package to screen for overlapping genes among the upregulated and downregulated DEGs in both datasets. This analysis revealed 197 overlapping genes in the upregulated gene set (Figure 1C) and 419 overlapping genes in the downregulated gene set (Figure 1D). In the context of analyzing PPI networks with overlapping genes, the MCODE algorithm yielded a network comprising 42 nodes and 794 edges, as illustrated in Figure 1E. The selected genes underwent GO and KEGG functional enrichment analysis, shedding light on their biological relevance. These genes enriched KEGG pathways including cell cycle, oocyte meiosis, and DNA replication (Figure 1F). Additionally, enrichment analysis unveiled significant associations with terms such as spindle assembly checkpoint signaling (BP), spindle, nucleus, CMG complex (CC), and single-stranded DNA helicase activity, protein kinase binding, kinase binding (MF) (Figure 1G).
Identification of seven signature prognostic genes associated with NSCLC
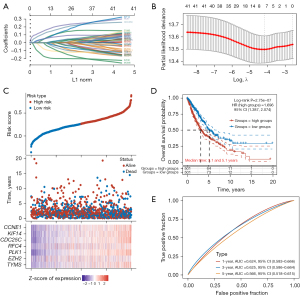
We used LASSO Cox regression analysis on 42 genes in the PPI network to look for possible predictive biomarkers. With the optimal lambda value λmin =0.0129 obtained by minimum likelihood deviation, seven important signature prognostic genes were obtained, namely EZH2, CDC25C, TYMS, RFC4, KIF14, PLK1, and CCNE1 (Figure 2A,2B). Individual risk scores were employed to divide NSCLC patients into two risk groups. Interestingly, patients in the high-risk group exhibited lower survival time and poorer outcomes compared to those in the low-risk group. Additionally, there were significant differences in the expression of seven genes among samples with different risk levels (Figure 2C). Secondly, the high-risk group had a lower OS probability, suggestive of a worse prognosis, according to the KM survival curve data (Figure 2D). The accuracy of the risk model in prognostic prediction was highlighted by the ROC curve analysis, which revealed that it had the greatest AUC value for 3-year prediction (Figure 2E).
CDC25C identified as independent prognostic factor in NSCLC
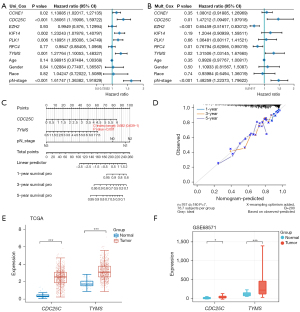
Univariate Cox regression analysis identified CDC25C and TYMS as significant prognostic markers, respectively (Figure 3A). In addition, multivariate Cox regression analysis identified EZH2 as an independent prognostic factor for NSCLC as well (Figure 3B). Predictive nomograms of 1-, 3-, and 5-year OS rates were generated (Figure 3C). The model exhibited strong performance in predicting OS at 1, 3, and 5 years, as shown by calibration curves that compared expected and actual survival outcomes (Figure 3D). We examined the expression of CDC25C and TYMS in NSCLC. When comparing the tumor group to the control group in the TCGA-NSCLC dataset, CDC25C and TYMS displayed greater expression (Figure 3E). Similarly, in the GSE68571 dataset, both CDC25C and TYMS showed elevated expression in the tumor group (Figure 3F). In this study, we selected CDC25C as the prognostic hub gene.
Knockdown of CDC25C inhibits NSCLC cell growth in vitro
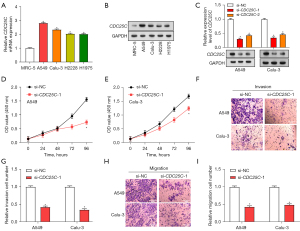
Expression of CDC25C in normal lung cells (MRC-5) and NSCLC cell lines (H1975, Calu-3, A549, and H2228) was evaluated using qRT-PCR and WB analysis (Figure 4A,4B). Notably, CDC25C was upregulated in NSCLC cell lines, among which A549 and Calu-3 cells were selected for further study due to their apparent differential expression. We evaluated the efficacy of the CDC25C knockdown using qRT-PCR and WB after introducing the vector into A549 and Calu-3 cells (Figure 4C). Among the small interfering RNA (siRNA) constructs tested, si-CDC25C-1 showed the most efficient knockdown, prompting its selection for subsequent experiments. We then examined the effects of CDC25C knockdown on the phenotypes of NSCLC cells (Figure 4D-4I). Our findings demonstrated that, in comparison to the control group, CDC25C knockdown dramatically reduced NSCLC cell migration, invasion, and proliferation.
Differential expression of CDC25C significantly affects apoptosis and autophagy of NSCLC cells
A CDC25C overexpression plasmid was introduced into A549 and Calu-3 cells, and the efficiency of overexpression was assessed using qRT-PCR and WB analyses, confirming the successful transfection (Figure 5A,5B). The apoptotic state of A549 and Calu-3 cells after CDC25C overexpression or knockdown was examined using flow cytometry (Figure 5C,5D). Our findings revealed that knockdown of CDC25C significantly promoted apoptosis, whereas its overexpression inhibited this process. Upon CDC25C overexpression or knockdown in NSCLC cells, we investigated the protein expression of the apoptotic proteins Bax and Bcl-2 (Figure 5E-5G). The data suggested that CDC25C knockdown increased the expression of Bax and reduced the levels of Bcl-2 compared to the control. Conversely, overexpression of CDC25C had the opposite effect. WB studies showed that CDC25C knockdown resulted in decreased LC3 I and LC3 II protein levels and increased p62 levels, indicating inhibition of autophagy. In contrast, when CDC25C was overexpressed, the protein levels were opposite (Figure 5H-5J).
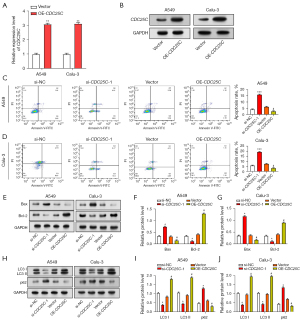
MiR-142-3p targets CDC25C and regulates NSCLC cell cycle
To investigate the upstream regulatory mechanism of CDC25C further, we predicted possible binding sites for CDC25C regulation using the StarBase database (http://starbase.sysu.edu.cn/), and found that the CDC25C 3'-UTR and miR-142-3p interacted (Figure 6A). The results of dual-luciferase experiments showed that the miR-142-3p mimic significantly reduced WT luciferase activity compared with the control group but did not significantly affect MUT (Figure 6B). In qRT-PCR experiments, compared with control cells, miR-142-3p was low-expressed in NSCLC cell lines (Figure 6C). Among them, A549 and Calu-3 showed the most significant differential expression and were selected for subsequent experiments. qRT-PCR measured the transfection efficiency of miR-142-3p in NSCLC cells (Figure 6D,6E). In contrast to the inhibitor of miR-142-3p, which produced low expression, the miR-142-3p mimic showed high expression in NSCLC cells. CCK-8 experiments also showed that low-expression miR-142-3p promoted cell proliferation in NSCLC, whereas miR-142-3p mimics inhibited proliferation (Figure 6F-6I). Similarly, the miR-142-3p mimic was found to promote the increase of NSCLC cells in the G2 phase (Figure 6J-6M). When the miR-142-3p overexpression model was introduced into cells, subsequent WB analysis revealed decreased levels of CDK1 and cyclin B1 protein and elevated levels of CDK1-Y15 phosphorylation (Figure 6N).
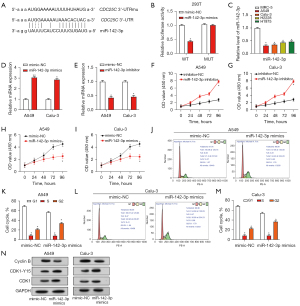
CDC25C reverses the inhibitory effect of miR-142-3p on cell cycle and proliferation of NSCLC cells
The introduction of the miR-142-3p inhibitor led to an elevation in the expression of CDC25C, whereas the introduction of the miR-142-3p mimic resulted in a reduction in the expression of CDC25C. This was demonstrated through both qRT-PCR and WB analysis (Figure 7A-7C). We next transfected the CDC25C overexpression plasmid and the miR-142-3p mimic, either separately or together, into A549 and Calu-3 cells. Increased expression of miR-142-3p was observed in cells after introduction of miR-142-3p mimics. qRT-PCR detection, however, revealed a minor decrease in the level of miR-142-3p as a consequence of CDC25C overexpression. When CDC25C overexpression and miR-142-3p mimic were co-transfected, the expression of miR-142-3p was greater than CDC25C overexpression alone (Figure 7D,7E). Moreover, the introduction of the miR-142-3p mimic led to an increase in CDK1-Y15 and CDC25C and a decrease in cyclin B protein levels, as indicated by the results of Figure 7F. In contrast, overexpression of CDC25C alone reversed these protein trends. The change in CDK1 protein level was not obvious. In contrast to controls, the expression of these proteins was somewhat restored when CDC25C overexpression was combined with a miR-142-3p mimic. Furthermore, miR-142-3p mimics were shown to suppress cell proliferation activity in CCK-8 studies, but overexpression of CDC25C reversed this effect and boosted cell proliferation activity (Figure 7G,7H).
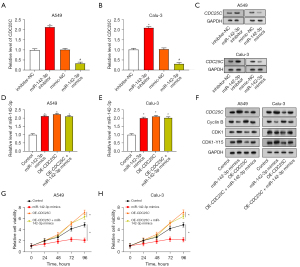
MiR-142-3p inhibition and CDC25C silencing regulate proliferation, apoptosis, and autophagy in NSCLC cells
From the results displayed in Figure 8A-8E, compared with the control, the miR-142-3p inhibitor increased NSCLC cell proliferation and reduced the apoptosis rate. Moreover, attenuating cell proliferation and enhancing apoptosis were achieved by the simultaneous downregulation of CDC25C and inhibition of miR-142-3p. After co-transfection of NSCLC cells with si-CDC25C-1 and miR-142-3p inhibitor, WB analysis results confirmed that the miR-142-3p inhibitor caused an increase in Bcl-2 levels and a decrease in Bax expression. Addition of si-CDC25C-1 reversed these effects (Figure 8F-8H). Moreover, LC3 I and LC3 II levels were elevated and p62 levels were downregulated by the miR-142-3p inhibitor, whereas si-CDC25C-1 introduction had the reverse result (Figure 8I-8K). This suggests that miR-142-3p might be able to regulate the pathways involved in the processes of autophagy and apoptosis by targeting CDC25C.
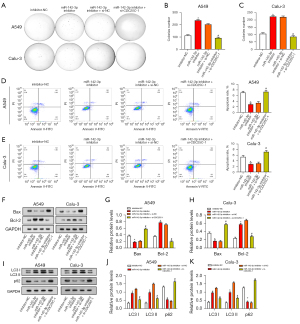
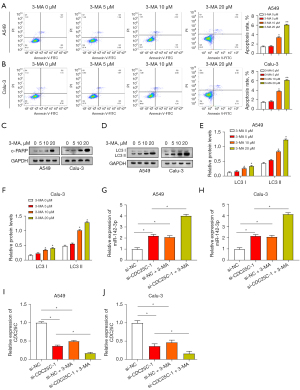
Autophagy inhibitor 3-MA promotes NSCLC cell apoptosis and regulates miR-142-3p/CDC25C expression
We used 3-MA (a specific autophagy inhibitor) to study the interaction between apoptosis and autophagy. A549 and Calu-3 cells were treated with 3-MA, and their apoptotic status was examined by flow cytometry (Figure 9A,9B). It is important to note that NSCLC cells induce apoptosis in response to varying doses of 3-MA and that this apoptotic impact intensifies as 3-MA concentration rises. These findings suggest that in NSCLC cells, autophagy suppression causes apoptosis. The expression of endogenous apoptosis marker c-PARP in NSCLC cells was subsequently analyzed (Figure 9C). As the 3-MA concentration increases, the expression of c-PARP also increases. WB analysis showed a positive association between the concentration of 3-MA and the levels of autophagy proteins. The change was particularly obvious at the concentration of 20 µM, so this concentration was selected for subsequent experiments (Figure 9D-9F). The expression levels of miR-142-3p and CDC25C were evaluated through qRT-PCR in A549 and Calu-3 cells subjected to CDC25C knockdown, along with the addition of 20 µM 3-MA. This experiment aimed to elucidate the involvement of the miR-142-3p/CDC25C axis in the regulation of autophagy. It was shown that the knockdown of CDC25C led to downregulation of CDC25C and upregulation of miR-142-3p. MiR-142-3p was more highly expressed following the co-knockdown of CDC25C and 3-MA, but CDC25C expression was further decreased (Figure 9G-9J). The above findings indicate that autophagy regulation is related to the miR-142-3p/CDC25C axis.
Discussion
NSCLC, the most prevalent and significant subtype of lung cancer, has attracted widespread attention in recent years (23). Due to its genetic variety and complexity, targeting NSCLC for therapeutic development has been proven difficult (24). Furthermore, the complex interactions between genetic changes and signaling pathways highlight the necessity of using customized medicine techniques in treatment plans. Through the selection and enrichment analysis of DEGs associated with NSCLC, we identified their close correlation with cellular processes including DNA replication and cell cycle. Existing research has elucidated that the deubiquitinase USP5 in NSCLC facilitates cell proliferation by physically binding to cell cycle protein D1 (CCND1), reducing its polyubiquitination levels, thereby enhancing cell cycle progression and tumorigenesis (25). A new study has also indicated that the long noncoding RNA SH3PXD2A-AS1, through interaction with DHX9, promotes NSCLC cell proliferation, accelerates the cell cycle, and amplifies FOXM1 expression (26). Cryptotanshinone (CTT) is a therapeutic agent for NSCLC that works by inhibiting the PI3K/Akt/GSK-3β pathway, which arrests the G0/G1 cell cycle and induces apoptosis (27). The development of NSCLC and cell cycle regulation are strongly correlated, according to all of these data.
In our study, CDC25C emerged as a robust prognostic gene in NSCLC, exhibiting superior predictive performance in prognosis models. The intricate regulation of CDC25C involves post-translational modifications, interactions with regulatory partners, and cell cycle-dependent degradation. Its association with various cancer types, including skin (28), cervical (29), and lung cancers (30), underscores its potential as a therapeutic target across diverse malignancies. Prior research has highlighted the critical role of genetic variations, particularly single nucleotide polymorphisms (SNPs) within the CDC25C family, as key factors influencing the survival and chemotherapy response in advanced NSCLC patients (31). Moreover, the selective splicing of the CDC25C gene has been identified as a crucial mechanism controlling the progression of the mitotic cell cycle and the development of lung cancer. MiRNAs, in particular those from the let-7 family, have been shown to regulate CDC25C, which affects how NSCLC cells react to different therapies (32). Together, these findings highlight the significant contribution and prognostic significance of CDC25C in NSCLC. Research has demonstrated that the upregulation of TRIM59 in NSCLC significantly influences the expression of cell cycle protein CDC25C, promoting proliferation and migration (33). Additionally, emerging studies on compounds such as 5-AcTMF (34), curcumin (35), and apatinib (36) have shed light on their potential to induce autophagy, and apoptosis, and inhibit NSCLC progression through distinct molecular pathways. These varied findings highlight the complex interactions between autophagy, cell cycle control, and possible therapeutic interventions in NSCLC, providing important information for the creation of tailored treatments. Furthermore, therapy response of patients with ‘low miR-142-3p’ to dimethyl fumarate (DMF), an established disease-modifying treatment (DMT), was superior to that of patients with ‘high miR-142-3p’ levels (37). In addition, study had shown that by screening and analyzing data from the CTRP and PRISM databases, the potential target gene CDC25C has been identified as being linked to the drugs oligomycin A and panobinostat, providing potential therapeutic options for treating patients with high-risk breast cancer (38). Our in vitro studies on the function of CDC25C in NSCLC cells showed that CDC25C downregulation efficiently inhibited NSCLC cell invasion, migration, and proliferation. Intriguingly, both knockdown and overexpression of CDC25C were found to exert a significant impact on the autophagic and apoptotic processes in NSCLC cells. The complex function of CDC25C in coordinating cellular responses in NSCLC is highlighted by these dual impacts on cell survival and programmed cell death.
To delve further into the specific mechanisms of CDC25C in NSCLC, we explored potential binding sites and identified an interaction with miR-142-3p. Interestingly, miR-142-3p is a miRNA that may attach to the mRNA of target genes and affect gene expression because of its distinct structural characteristics (39,40). Its function as a tumor suppressor (41) or promoter (42) via targeting several genes has been clarified by extensive investigation. Research indicates that overexpression of miR-142-3p in NSCLC has been shown to increase chemotherapy sensitivity by blocking HMGB1-mediated autophagy (43). This underscores its potential therapeutic significance for NSCLC patients. Through the miR-142-3p-HMGB1 axis, the downregulation of circ-YES1 in NSCLC has been connected to the suppression of cell migration and proliferation, suggesting a possible therapeutic target for NSCLC therapy (44). Moreover, miR-142-3p functions as an anti-cancer agent in NSCLC by suppressing the expression of NR2F6, preventing cell division, and triggering apoptosis in NSCLC tissues and cell lines (45). This finding raises the possibility of a novel approach to overcome treatment resistance in NSCLC. These findings collectively emphasized the intricate regulatory network involving CDC25C and miR-142-3p in NSCLC pathogenesis and treatment, providing valuable insights for developing targeted therapeutic approaches. Furthermore, via blocking autophagy, miR-142-3p overexpression was associated with increased chemotherapy sensitivity, indicating a possible method to combat drug resistance in the treatment of NSCLC.
As part of our investigation into the molecular dynamics inside NSCLC, we carefully analyzed the proteins that regulate cell cycle regulation, proliferation, apoptosis, and autophagy to shed light on the complicated roles of miR-142-3p and CDC25C. Cell cycle proteins critical for cell cycle progression were studied to assess cell proliferation (46). Notably, altered CDK1 phosphorylation patterns at Y15 and overexpression of cyclin B, a crucial driver of the G2 phase transition, suggested a breakdown in cell cycle control (47). These variations in cyclin B levels have the potential to trigger an early commencement of mitosis, which might result in chromosomal instability and atypical cell division, ultimately enabling carcinogenic processes (48,49). Additionally, research on the proteins Bax and Bcl-2, which are pro- and anti-apoptotic, has provided insight into the intricate balance that controls decisions about cellular fate (50,51). Bax expression or functionality changes may underpin apoptosis resistance pathways, impacting NSCLC cell survival. We assessed autophagy by measuring the amounts of LC3 I and LC3 II, which are required for autophagosome formation, and p62, a selective substrate for autophagic breakdown. Both p62 accumulation and the transition from LC3 I to LC3 II functioned as biomarkers for autophagy activation and autophagic turnover, respectively, and are important markers of autophagic activity in the cellular setting of NSCLC (52-54). Our research highlighted that miR-142-3p plays a pivotal role in the regulation of NSCLC, exerting its influence on the cell cycle by targeting key components such as cyclin B, CDK1, and CDK1-Y15. Cell proliferation is promoted by upregulating CDC25C levels through inhibition of miR-142-3p. MiR-142-3p mimics, on the other hand, caused cell cycle arrest and decreased proliferation. Furthermore, miR-142-3p also regulates apoptosis- and autophagy-related proteins, emphasizing its potential as a treatment for NSCLC.
We investigated the impact of autophagy suppression on cell function experimentally by using 3-MA, an autophagy inhibitor, to prevent the development of autophagosomes. In NSCLC, for example, the chemical CA-5f has strong anti-tumor effects via increasing the production of reactive oxygen species (ROS) produced from mitochondria, causing cell apoptosis, and suppressing autophagic flux (55). Furthermore, investigations into autophagy inhibition have extended to combinations of agents, such as the AS-IV/propofol combination, where the autophagy inhibitor 3-MA has been shown to enhance its pro-apoptotic capabilities in NSCLC cells (56). This suggests a potential tumor-promoting role of autophagy in NSCLC, emphasizing the need for targeted interventions to modulate autophagic processes in cancer therapy. Moreover, a study had demonstrated that the autophagy inhibitor 3-MA can reduce platinum resistance in NSCLC cells and promote apoptosis induced by platinum-based chemotherapy (57). Our study revealed that NSCLC cells treated with the autophagy inhibitor 3-MA saw a dose-dependent increase in apoptosis, suggesting that autophagy plays a crucial role in cell survival. Overexpression of the apoptosis marker c-PARP and increased LC3 I and LC3 II protein levels associated with autophagy inhibition highlight the potential of 3-MA as a therapeutic agent for NSCLC. In addition, the way that 3-MA treatment regulates the miR-142-3p/CDC25C axis implies its involvement in autophagy regulation, providing insights into a novel regulatory mechanism for NSCLC tumor progression. We chose NSCLC as the subject of our study to expand the understanding of the role of miR-142-3p in different tumor areas. Further insights into the molecular details of miR-142-3p inhibition of cancer cell proliferation via CDC25C were provided. More detailed experimental evidence was provided to reveal the specific pathways and molecular interactions of miR-142-3p. And a series of different experimental tools were used to validate the results and exclude other potential influences to ensure a comprehensive and in-depth understanding of the role of miR-142-3p. However, there are some limitations in our study. Although NSCLC was selected as the study population, there may be other subtypes of lung cancer that were not considered, and therefore the results may not be generalizable. Although the mechanism of miR-142-3p has been thoroughly investigated, there may still be factors that have not been taken into account to influence the results, such as cellular microenvironment and individual differences. In addition, in vivo experiments have not been performed to confirm these findings.
Conclusions
To summarize, CDC25C has a likely useful predictive value and is strongly linked to important processes such as the cell cycle, according to our thorough research and screening of NSCLC-related genes. The control of CDC25C expression by knockdown and overexpression revealed the regulation of autophagy, apoptosis, invasion and proliferation of cells. The CDC25C/miR-142-3p axis had a substantial impact on autophagy and apoptosis in NSCLC cells, as evidenced by additional investigation into the interaction between the two factors. These results may open the door for more focused treatment approaches in the future and further our knowledge of the intricate molecular mechanisms behind NSCLC development.
Acknowledgments
We would like to extend our heartfelt thanks to the editorial experts for their meticulous assistance and invaluable suggestions for the improvement of this manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD and MDAR checklists. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-82/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-82/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-82/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-82/coif). The authors report that this work was supported by the Minsheng Research Project of Pudong New Area Science and Technology Development Fund (No. PKJ2023-Y15), the Natural Science Fund of Shanghai (No. 21ZR1411200), the National Natural Science Foundation of China (No. 82272768), the Natural Science Fund of Beijing (No. 236678), and the Medical Discipline Construction Project of Pudong Health Committee of Shanghai (No. PWYts2021-18). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Inage T, Nakajima T, Yoshino I, et al. Early Lung Cancer Detection. Clin Chest Med 2018;39:45-55. [Crossref] [PubMed]
- Wang S, Zimmermann S, Parikh K, et al. Current Diagnosis and Management of Small-Cell Lung Cancer. Mayo Clin Proc 2019;94:1599-622. [Crossref] [PubMed]
- Kaur R, Suresh PK. Chemoresistance Mechanisms in Non-Small Cell Lung Cancer-Opportunities for Drug Repurposing. Appl Biochem Biotechnol 2023; Epub ahead of print. [Crossref]
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Wagland R, Brindle L, James E, et al. Facilitating early diagnosis of lung cancer amongst primary care patients: The views of GPs. Eur J Cancer Care (Engl) 2017;26:e12704. [Crossref] [PubMed]
- Araghi M, Mannani R, Heidarnejad Maleki A, et al. Recent advances in non-small cell lung cancer targeted therapy; an update review. Cancer Cell Int 2023;23:162. [Crossref] [PubMed]
- Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med 2021;27:1345-56. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Stern E, Pines G, Lazar LO, et al. CDC25C Protein Expression Correlates with Tumor Differentiation and Clinical Outcomes in Lung Adenocarcinoma. Biomedicines 2023;11:362. [Crossref] [PubMed]
- Li FN, Zhang QY, Li O, et al. ESRRA promotes gastric cancer development by regulating the CDC25C/CDK1/CyclinB1 pathway via DSN1. Int J Biol Sci 2021;17:1909-24. [Crossref] [PubMed]
- Xu N, Ren Y, Bao Y, et al. PUF60 promotes cell cycle and lung cancer progression by regulating alternative splicing of CDC25C. Cell Rep 2023;42:113041. [Crossref] [PubMed]
- Liao CC, Chen SC, Huang HP, et al. Gallic acid inhibits bladder cancer cell proliferation and migration via regulating fatty acid synthase (FAS). J Food Drug Anal 2018;26:620-7. [Crossref] [PubMed]
- Li L, Zou BJ, Zhao JZ, et al. A novel DNA damage repair-related signature for predicting prognositc and treatment response in non-small lung cancer. Front Oncol 2022;12:961274. [Crossref] [PubMed]
- Chatterjee S, Huang EH, Christie I, et al. Reactivation of the p90RSK-CDC25C Pathway Leads to Bypass of the Ganetespib-Induced G2-M Arrest and Mediates Acquired Resistance to Ganetespib in KRAS-Mutant NSCLC. Mol Cancer Ther 2017;16:1658-68. [Crossref] [PubMed]
- Xue T, Chen Y, Xu J, et al. Cyclovirobuxine D inhibits growth and progression of non-small cell lung cancer cells by suppressing the KIF11-CDC25C-CDK1-CyclinB1 G(2)/M phase transition regulatory network and the NFκB/JNK signaling pathway. Int J Oncol 2023;62:57. [Crossref] [PubMed]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [Crossref] [PubMed]
- Goh JN, Loo SY, Datta A, et al. microRNAs in breast cancer: regulatory roles governing the hallmarks of cancer. Biol Rev Camb Philos Soc 2016;91:409-28. [Crossref] [PubMed]
- Schmidt L, Fredsøe J, Kristensen H, et al. Training and validation of a novel 4-miRNA ratio model (MiCaP) for prediction of postoperative outcome in prostate cancer patients. Ann Oncol 2018;29:2003-9. [Crossref] [PubMed]
- Duran-Sanchon S, Moreno L, Augé JM, et al. Identification and Validation of MicroRNA Profiles in Fecal Samples for Detection of Colorectal Cancer. Gastroenterology 2020;158:947-957.e4. [Crossref] [PubMed]
- Zhu K, Zhang Z, Zhang H, et al. MiR-142-3p targeting NUCKS1 inhibits proliferation and invasion of pancreatic cancer cells. Artif Cells Nanomed Biotechnol 2020;48:415-24. [Crossref] [PubMed]
- Qi X, Li J, Zhou C, et al. MiR-142-3p Suppresses SOCS6 Expression and Promotes Cell Proliferation in Nasopharyngeal Carcinoma. Cell Physiol Biochem 2015;36:1743-52. [Crossref] [PubMed]
- Lachowicz D, Mielczarek P, Wirecka R, et al. Nanohydrogels Based on Self-Assembly of Cationic Pullulan and Anionic Dextran Derivatives for Efficient Delivery of Piroxicam. Pharmaceutics 2019;11:622. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Liu WJ, Du Y, Wen R, et al. Drug resistance to targeted therapeutic strategies in non-small cell lung cancer. Pharmacol Ther 2020;206:107438. [Crossref] [PubMed]
- Zhang Z, Cui Z, Xie Z, et al. Deubiquitinase USP5 promotes non-small cell lung cancer cell proliferation by stabilizing cyclin D1. Transl Lung Cancer Res 2021;10:3995-4011. [Crossref] [PubMed]
- Zhou Y, Yong H, Cui W, et al. Long noncoding RNA SH3PXD2A-AS1 promotes NSCLC proliferation and accelerates cell cycle progression by interacting with DHX9. Cell Death Discov 2022;8:192. [Crossref] [PubMed]
- Kim SA, Kang OH, Kwon DY. Cryptotanshinone Induces Cell Cycle Arrest and Apoptosis of NSCLC Cells through the PI3K/Akt/GSK-3β Pathway. Int J Mol Sci 2018;19:2739. [Crossref] [PubMed]
- Al-Matouq J, Holmes TR, Hansen LA. CDC25B and CDC25C overexpression in nonmelanoma skin cancer suppresses cell death. Mol Carcinog 2019;58:1691-700. [Crossref] [PubMed]
- Ruan X, Jiang J. RACGAP1 promotes proliferation and cell cycle progression by regulating CDC25C in cervical cancer cells. Tissue Cell 2022;76:101804. [Crossref] [PubMed]
- Zhang W, Shang X, Yang F, et al. CDC25C as a Predictive Biomarker for Immune Checkpoint Inhibitors in Patients With Lung Adenocarcinoma. Front Oncol 2022;12:867788. [Crossref] [PubMed]
- Zheng Y, Deng Z, Tang M, et al. Impact of genetic factors on platinum-induced gastrointestinal toxicity. Mutat Res Rev Mutat Res 2020;786:108324. [Crossref] [PubMed]
- Yu H, Xu L, Liu Z, et al. Circ_MDM2_000139, Circ_ATF2_001418, Circ_CDC25C_002079, and Circ_BIRC6_001271 Are Involved in the Functions of XAV939 in Non-Small Cell Lung Cancer. Can Respir J 2019;2019:9107806. [Crossref] [PubMed]
- Zhan W, Han T, Zhang C, et al. TRIM59 Promotes the Proliferation and Migration of Non-Small Cell Lung Cancer Cells by Upregulating Cell Cycle Related Proteins. PLoS One 2015;10:e0142596. [Crossref] [PubMed]
- Li YR, Li S, Ho CT, et al. Tangeretin derivative, 5-acetyloxy-6,7,8,4'-tetramethoxyflavone induces G2/M arrest, apoptosis and autophagy in human non-small cell lung cancer cells in vitro and in vivo. Cancer Biol Ther 2016;17:48-64. [Crossref] [PubMed]
- Xie H, Huang G, Zou J, et al. The Pharmacological Mechanism of Curcumin against Drug Resistance in Non-Small Cell Lung Cancer: Findings of Network Pharmacology and Bioinformatics Analysis. Evid Based Complement Alternat Med 2022;2022:5926609. [Crossref] [PubMed]
- Xie C, Zhou X, Liang C, et al. Apatinib triggers autophagic and apoptotic cell death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/p62 signaling in lung cancer. J Exp Clin Cancer Res 2021;40:266. [Crossref] [PubMed]
- De Vito F, Musella A, Fresegna D, et al. MiR-142-3p regulates synaptopathy-driven disease progression in multiple sclerosis. Neuropathol Appl Neurobiol 2022;48:e12765. [Crossref] [PubMed]
- Tian S, Fu L, Zhang J, et al. Identification of a DNA Methylation-Driven Genes-Based Prognostic Model and Drug Targets in Breast Cancer: In silico Screening of Therapeutic Compounds and in vitro Characterization. Front Immunol 2021;12:761326. [Crossref] [PubMed]
- Zheng J, Cheng C, Xu J, et al. miR-142-3p Regulates Tumor Cell Autophagy and Promotes Colon Cancer Progression by Targeting TP53INP2. Chemotherapy 2022;67:57-66. [Crossref] [PubMed]
- Ren J, Li W, Pan G, et al. miR-142-3p Modulates Cell Invasion and Migration via PKM2-Mediated Aerobic Glycolysis in Colorectal Cancer. Anal Cell Pathol (Amst) 2021;2021:9927720. [Crossref] [PubMed]
- Mansoori B, Duijf PHG, Mohammadi A, et al. MiR-142-3p targets HMGA2 and suppresses breast cancer malignancy. Life Sci 2021;276:119431. [Crossref] [PubMed]
- Hu Z, Yin Y, Jiang J, et al. Exosomal miR-142-3p secreted by hepatitis B virus (HBV)-hepatocellular carcinoma (HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression. J Gastrointest Oncol 2022;13:754-67. [Crossref] [PubMed]
- Cui Y, Wu X, Jin J, et al. CircHERC1 promotes non-small cell lung cancer cell progression by sequestering FOXO1 in the cytoplasm and regulating the miR-142-3p-HMGB1 axis. Mol Cancer 2023;22:179. [Crossref] [PubMed]
- Jin M, Wang Y, Zhou D, et al. Downregulation of circ-YES1 suppresses NSCLC migration and proliferation through the miR-142-3p-HMGB1 axis. Respir Res 2023;24:100. [Crossref] [PubMed]
- Jin C, Xiao L, Zhou Z, et al. MiR-142-3p suppresses the proliferation, migration and invasion through inhibition of NR2F6 in lung adenocarcinoma. Hum Cell 2019;32:437-46. [Crossref] [PubMed]
- Sun J, Wang X, Shen Q, et al. DNASE1L3 inhibits hepatocellular carcinoma by delaying cell cycle progression through CDK2. Cell Oncol (Dordr) 2022;45:1187-202. [Crossref] [PubMed]
- Xie B, Wang S, Jiang N, et al. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett 2019;443:56-66. [Crossref] [PubMed]
- Barbiero M, Cirillo L, Veerapathiran S, et al. Cell cycle-dependent binding between Cyclin B1 and Cdk1 revealed by time-resolved fluorescence correlation spectroscopy. Open Biol 2022;12:220057. [Crossref] [PubMed]
- Deota S, Rathnachalam S, Namrata K, et al. Allosteric Regulation of Cyclin-B Binding by the Charge State of Catalytic Lysine in CDK1 Is Essential for Cell-Cycle Progression. J Mol Biol 2019;431:2127-42. [Crossref] [PubMed]
- Lopez A, Reyna DE, Gitego N, et al. Co-targeting of BAX and BCL-XL proteins broadly overcomes resistance to apoptosis in cancer. Nat Commun 2022;13:1199. [Crossref] [PubMed]
- Brayer S, Joannes A, Jaillet M, et al. The pro-apoptotic BAX protein influences cell growth and differentiation from the nucleus in healthy interphasic cells. Cell Cycle 2017;16:2108-18. [Crossref] [PubMed]
- Xin X, Wu M, Meng Q, et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer 2018;17:94. [Crossref] [PubMed]
- Peña-Martinez C, Rickman AD, Heckmann BL. Beyond autophagy: LC3-associated phagocytosis and endocytosis. Sci Adv 2022;8:eabn1702.
- Heckmann BL, Green DR. LC3-associated phagocytosis at a glance. J Cell Sci 2019;132:jcs222984. [Crossref] [PubMed]
- Zhang L, Qiang P, Yu J, et al. Identification of compound CA-5f as a novel late-stage autophagy inhibitor with potent anti-tumor effect against non-small cell lung cancer. Autophagy 2019;15:391-406. [Crossref] [PubMed]
- Liu J, Chen L, Zhang J, et al. AS-IV enhances the antitumor effects of propofol in NSCLC cells by inhibiting autophagy. Open Med (Wars) 2023;18:20230799. [Crossref] [PubMed]
- Li Z, Wang Y, Wu L, et al. Apurinic endonuclease 1 promotes the cisplatin resistance of lung cancer cells by inducing Parkin-mediated mitophagy. Oncol Rep 2019;42:2245-54. [Crossref] [PubMed]


