
Differential expression of DMD transcripts as a novel prognostic biomarker in histologically diverse mesotheliomas
Highlight box
Key findings
• Expression of the Duchenne muscular dystrophy (DMD) gene in primary mesothelioma was associated with a 2-year reduction in survival for patients with high transcript levels. In a multivariate model, DMD expression predicted overall survival across all histological subtypes independently of genetic alterations previously linked to poor mesothelioma prognosis.
What is known and what is new?
• The 5-year mesothelioma survival is poor. A prognostically-relevant molecular marker to guide the clinical management is urgently needed. A role for the DMD gene expression as a predictor of survival across various tumours has been demonstrated recently.
• In mesothelioma, differential expression of specific transcripts from this largest human gene known was associated with patients’ survival. High expression corresponded with poor survival. The genes differentially expressed between the high versus low expression groups revealed alterations in specific pathways relevant to cancer progression.
What is the implication, and what should change now?
• Expression of specific DMD transcript identifies patients at risk of poor survival, and therefore should be explored as a predictive biomarker to inform treatment intensity, follow-up schedules, eligibility for clinical trials, and end-of-life care planning.
Introduction
Background
The incidence of mesothelioma is varied across countries, with Northern Europe reporting the highest rates (0.30 per 100,000), and males having significantly higher rates and worse survival than females (1). Although these rates are in decline due to the elimination of asbestos, mesothelioma has a latency period of 30 to 40 years and considerable amounts of asbestos are still being utilised in developing countries (1). Furthermore, despite that substantial advances have been made in understanding the molecular biology of mesothelioma (2), the molecular basis of this disease is not yet fully understood (3).
The majority of patients are diagnosed with unresectable or advanced stage disease, and they receive platinum-based chemotherapy or, more recently, combination immunotherapy as the first-line treatment (4). Nevertheless, mesothelioma invariably leads to death, and the 5-year survival rate remains at 12% (1). Therefore, it remains a significant clinical challenge.
The three major histological subtypes of mesothelioma are epithelioid, sarcomatoid and biphasic, with the epithelioid type having the most favourable prognosis (5). This classification has been challenged by genomic and transcriptomic analyses, which revealed extensive interpatient heterogeneity (6) that is further exacerbated by inflammation, cellularity and vacuolisation within the stroma surrounding the tumour (7).
This molecular heterogeneity and rare long-term survival have hindered the identification of prognostically relevant molecular markers, which are urgently needed to enhance our understanding of the biology of this disease, guide the clinical management, and potentially uncover novel therapeutic targets (5,8).
The most reliable predictors of survival continue to be the clinical and pathological parameters (8). Besides histology, which is still considered the most accurate prognostic factor, clinical prognostic indicators include age, disease stage, haemoglobin levels, white blood cell and platelet counts, performance status, and lactate dehydrogenase (LDH) levels (5).
Many studies have explored predictive molecular markers for mesothelioma, but none found routine clinical application [reviewed in the study (5)]. Homozygous deletions of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene, which are found in 60% to 73% of mesothelioma patients, have been linked to poor mesothelioma prognosis (9,10). However, these deletions have not been demonstrated to outperform the standard clinicopathological parameters in predicting mesothelioma survival (5,8,9). Another frequently mutated gene in mesothelioma is neurofibromatosis type 2 (NF2) with 30–40% of mesotheliomas harbouring NF2 somatic mutations (11,12). The association of hemizygous NF2 deletions with worse mesothelioma prognosis is not unequivocal (13-15). Finally, inactivating mutations in the BRCA1-associated protein-1 (BAP1) gene have been detected in up to 67% of mesotheliomas (16,17). While some studies have linked BAP1 protein loss to improved mesothelioma prognosis (16,18), others found no impact on survival (14,15). The low sensitivity and technical complexity (e.g., fluorescence in situ hybridisation) of the detection methods of these markers restricts their widespread clinical use (14,17).
A role for the Duchenne muscular dystrophy (DMD) gene expression as a predictor of survival across various tumours has been highlighted, including different carcinomas, haematological malignancies, and low-grade gliomas (19-22). The DMD gene is the second largest human gene known, with 79 exons and eight independent tissue-specific promoters (23). Three promoters control the expression of 14-kb full-length transcripts encoding 427 kDa dystrophins (Dp427). Five intragenic promoters give rise to transcripts encoding truncated isoforms, of which Dp71 is the most ubiquitous (23). Alternative splicing adds further structural and functional diversity (Figure 1). The main Dp71 splice variants (24) result from alternative splicing events involving exons 71 and 78. Dp71a lacks exon 71, Dp71b lacks exon 78, and Dp71ab lacks both exons 71 and 78. The absence of exon 78 leads to the replacement of the hydrophilic C-terminus with a unique hydrophobic one (24).

Notably, Duchenne abnormalities, including dysregulation of various signalling pathways (25), asymmetric cell division (26), and alterations in cell proliferation and migration (27,28) are also typical for malignancy and, indeed, have been seen in myogenic tumours (28).
Importantly, the reduction of DMD gene expression in tumours compared to control tissues was due to transcriptional downregulation and not somatic mutations, which could be expected in malignancies (20). This specific regulation indicates the importance of this mechanism.
Rationale and knowledge gap
The molecular heterogeneity and short survival have hindered the identification of prognostically relevant mesothelioma biomarkers, which could guide the clinical management and help identify new therapeutic targets. Interestingly, a comprehensive analysis of specific DMD gene transcripts revealed a significant complexity suggesting that individual variants may have distinct roles in tumours. While the transcript encoding the full-length muscle specific isoform (Dp427m) was downregulated across primary tumours, the transcripts encoding Dp71 variants showed variability of expression (20).
Objective
Given these findings, we performed a targeted analysis of DMD transcripts in various mesothelioma types and examined the association between their expression and patient survival. Next, we investigated Dp71 as a unique biomarker across mesotheliomas. We present this article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-28/rc).
Methods
Data acquisition
Normalised and log transformed RNAseq expression data for the DMD gene in primary mesothelioma samples and corresponding clinical data, including the demographics of patients, the histological subtype of samples and tumour stage, were obtained from the University of California Santa Cruz (UCSC) Xena Functional Genomics Browser (http://xena.ucsc.edu) after selecting The Cancer Genome Atlas (TCGA) Mesothelioma (MESO) cohort and querying for the DMD gene. Somatic mutation data were obtained from cBioPortal (https://www.cbioportal.org). Gene-level copy number variation (CNV) data estimated using the Genomic Identification of Significant Targets in Cancer (GISTIC2.0) method were obtained from the XENA Browser (29,30).
The TCGA RNAseq isoform data were downloaded from the MESO mRNASeq archives “illuminahiseq_rnaseqv2-RSEM_isoforms_normalized MD5” at Broad Institute TCGA Genome Data Analysis Centre Firebrowse portal (http://firebrowse.org). UCSC identifiers of transcripts were matched to specific DMD transcripts using the UCSC genome browser (uc004dda.1: Dp427m; uc004dcm.1: Dp71; uc004dcn.1: Dp71a, uc004dco.1: Dp71b, and uc004dcp.1: Dp71ab). However, we note that estimates of transcript expression by RSEM may not be 100% accurate. Alternative splicing may result in RNAseq reads mapping to multiple transcripts of the same gene or multiple genes in different genomic locations (31) leading to mapping ambiguities (32).
Gene-level raw RNAseq expression data used for the differential gene expression analysis were also downloaded from the Firebrowse portal. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Comparisons of the DMD gene/transcripts expression levels
Mesothelioma samples were classified into two groups: epithelioid, and non-epithelioid mesothelioma [biphasic, sarcomatoid, and not otherwise specified (NOS) samples]. Data were tested for normality using a Shapiro-Wilk test in GraphPad Prism version 9.5.0 (San Diego, CA, USA) and homogeneity of variances using Levene’s test. Normally distributed data were analysed using an unpaired t-test or an ordinary one-way ANOVA (when groups had equal variances). Non-normally distributed data were analysed using a Mann-Whitney test or Kruskal-Wallis and Dunn’s multiple comparisons tests.
Cut-point determination
Datasets were divided into low and high DMD gene/transcript expression groups using the X-Tile software version 3.6.1 (Yale University School of Medicine, USA) (33). Optimal cut-off values were determined using a minimal P value approach. The software tests various expression thresholds, and the threshold that yields the smallest P value, signifying the most significant difference in outcomes between the two groups, is selected as the cut-off point.
The cut-point for the total DMD dataset was 7.19 log2(norm_count +1). The cut-points for Dp71, Dp71a, Dp71b, Dp71ab and Dp427m were 48.11, 0.01, 51.37, 105.09 and 2.57 normalised RSEM, respectively. The differences in expression between the high versus low groups for DMD gene/transcripts are shown in Figure S1.
Survival analysis
Kaplan-Meier curves were generated in GraphPad and the and statistical analysis of survival was performed using the log-rank test. Age at diagnosis data was split into three groups: patients below 60 years old, patients in their 60s, and aged 70 and above. When two or three comparisons were performed, the alpha value was adjusted to 0.025 or 0.017, respectively. For multivariate analyses, Cox regression analysis was conducted in SPSS (IBM Corp., Armonk, NY, USA).
Contingency tests
We used Fisher’s exact test to assess the presence of associations between high versus low DMD expression and the occurrence of DMD deletions and somatic mutations. The test was performed in GraphPad Prism.
Differential gene expression analysis
The differential gene expression analysis was performed using Limma Voom (34) in R v4.0.4 to identify the differentially expressed genes (DEGs) between the high versus low DMD gene/transcript expression groups. Genes with a false discovery rate (FDR)-corrected P value <0.05, and a |Log fold change (FC)| value ≥0.7 were considered differentially expressed. The EnrichR tool (35) was used to identify the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways enriched in the DEGs.
Correlation between DMD gene/transcripts expression and tumour microenvironment (TME) cells
The percentages of different stromal and immune cells [including natural killer (NK) and endothelial cells, macrophages, and fibroblasts] within the TME cells for mesothelioma samples predicted by the Kassandra algorithm (36) were downloaded from (https://science.bostongene.com/kassandra/api/download/TCGA_predicted_by_Kassandra.tar.gz). Pearson correlation between DMD gene/transcript expression and percentages of TME cells was calculated using R v4.0.4.
Results
Expression of DMD transcripts in mesothelioma samples
The TCGA MESO cohort used in this study included 87 mesothelioma samples with the following histological subtypes: 57 epithelioid, and 30 non-epithelioid (23 biphasic, two sarcomatoid, and five NOS). The age of patients at the initial diagnosis ranged between 28 and 81 years. Seventy-one of the patients were males and 16 were females. The pathologic stages of cases were stage I (n=10), stage II (n=16), stage III (n=45) and stage IV (n=16). One sample of the sarcomatoid subtype was not included in the survival analyses due to the lack of survival data.
We examined the expression of the full-length isoform (Dp427m) and splice variants of the Dp71 isoform in mesothelioma.
Dp427m, Dp71b and Dp71ab had the highest expression level in primary mesothelioma samples (Figure 2A). Dp71 and Dp71b expression was higher in non-epithelioid mesothelioma compared to the epithelioid subtype (P=0.04 and P=0.003, respectively; Figure 2B). In contrast, Dp427m expression was higher in epithelioid than non-epithelioid mesothelioma (P=0.04; Figure 2B).

To explore the interplay and regulatory relationships between these transcripts and to identify potential co-regulation or differential expression patterns, we evaluated the correlations between Dp71, Dp71a, Dp71b, Dp71ab, and Dp427m, and the total DMD gene expression (Figure 2C). We found a strong positive correlation between total DMD and Dp427m expression (R=0.73, P<0.001) and between total DMD and Dp71b expression (R=0.61, P<0.001). We also found a moderate positive correlation between total DMD and Dp71ab (R=0.58, P<0.001), Dp71 and Dp71a (R=0.48, P<0.001), and Dp71 and Dp71b (R=0.44, P<0.001). Finally, we found a weak positive correlation between Dp71ab and Dp427m expression (R=0.34, P=0.001), Dp71b and Dp427m (R=0.32, P=0.002), total DMD and Dp71 (R=0.27, P=0.01), and Dp71b and Dp71ab (R=0.26, P=0.01; Figure 2D).
Association between the expression of DMD gene/transcripts and survival
In a univariate analysis, total DMD gene expression had a significant effect on the overall survival (OS) of mesothelioma patients, with about a year difference in median survival time between the high versus low DMD groups. The median survival time for the high DMD group was 14.99 months compared to 27.16 months for the low DMD group (Figure 3A).
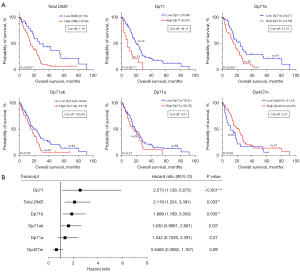
Likewise, high Dp71 transcript expression was associated with poor mesothelioma patient survival. Of the specific Dp71 splice variants, high Dp71b and high Dp71ab expression were also associated with poor survival (Figure 3A). The differences in median OS for the high versus low groups were 11.6, 9 and 3.8 months for Dp71, Dp71b, and Dp71ab, respectively. The median survival times were 8.351, 14.99, and 16.18 months for the high Dp71, Dp71b, and Dp71ab groups, respectively, compared to 19.99, 24.07, and 19.99 months for the low Dp71, Dp71b, and Dp71ab groups, respectively. The hazard ratios (HRs) and P values are displayed in Figure 3B.
No association between Dp71a or Dp427m expression and mesothelioma patient survival was found (Figure 3A). Given that Dp427m is highly expressed and Dp71a shows low levels in mesothelioma samples, this reinforces the association of other transcripts with survival.
As mutations or deletions could be responsible for the low DMD gene expression, the presence of somatic mutations and CNVs in high and low DMD groups was interrogated. We found no overrepresentation of shallow or deep deletions or mutations in the DMD locus in either high or low expression groups (P>0.99). In the low DMD group 33.3% of samples had DMD gene deletions or mutations compared to 35.5% in the high DMD group.
Pathways associated with the altered expression of DMD gene/transcripts
Pathway enrichment analysis in the DEGs between the high versus low DMD expression groups identified three pathways (Figure S2, Table S1): extracellular matrix (ECM)-receptor interaction (P=0.01), arrhythmogenic right ventricular cardiomyopathy (P=0.01), and focal adhesion (P=0.04).
The pathways enriched in the DEGs between high versus low Dp71 groups included cell cycle (P=3.01×10−4) and homologous recombination (P=0.01; Figure S2, Table S2). The pathways enriched between high versus low Dp71b groups included focal adhesion (P=5.94×10−6), ECM-receptor interaction (P=3.53×10−5), PI3K-Akt signalling (P=4.50×10−5), and calcium signalling (P=0.02; Figure S2, Table S3). Finally, the pathways enriched between high versus low Dp71ab groups included ECM-receptor interaction (P=1.78×10−4), PI3K-Akt signalling pathway (P=0.01), and focal adhesion (P=0.03; Figure S2, Table S4). This indicates that the Dp71b transcript encoding the alternative C-terminus is predominantly associated with cell adhesion, calcium and PI3K-Akt signalling.
No pathways were identified for the Dp71a or Dp427m comparison, which aligns with the lack of association between Dp427m or Dp71a expression and survival of mesothelioma patients.
Correlation between the expression of DMD gene/transcripts and TME cells
Given the importance of TME in tumour progression and response to treatment (37), we examined the correlation between the of DMD gene/transcripts expression and TME cells in the mesothelioma samples (Figure S3). We did this by using previously published predictions of the percentage of various cell types in the TME in MESO TCGA samples derived from gene expression data by deconvolution (36).
We found a weak negative correlation between DMD expression and the predicted percentage of NK cells (R=−0.24, P=0.02). We also found a weak positive correlation between Dp71 transcript expression and the predicted percentage of macrophages (R=0.32, P=0.002) and M2 macrophages (R=0.34, P=0.001), a weak positive correlation between Dp71b expression and the predicted percentage of endothelium (R=0.27, P=0.01) and fibroblasts (R=0.32, P=0.003), and a weak positive correlation between Dp71ab expression and the predicted percentage of fibroblasts (R=0.26, P=0.01).
Given that high expression levels of total DMD gene, Dp71, Dp71b, and Dp71ab transcripts were associated with worse survival (Figure 2A) and given the observed correlations between these transcripts in mesothelioma (Figure 2D), and their correlations with TME cells (Figure S3), we investigated whether the expression of pairs of these transcripts more effectively predict OS compared to each transcript alone. Patients were categorised into high/high (HH), high/low (HL), low/high (LH), and low/low (LL) groups and OS was compared. Patients in the HH total DMD/Dp71, HH total DMD/Dp71b, and HH total DMD/Dp71ab groups had worse survival than their corresponding LL groups (Figure 4A). The median survival times of patients in the HH total DMD/Dp71ab, HH total DMD/Dp71, and HH total DMD/Dp71b, groups were 27.79, 23.9, and 18.11 months shorter, respectively, than their corresponding LL groups.

Median survival times of patients in the HH Dp71/Dp71ab, HH Dp71/Dp71b and HH Dp71b/Dp71ab groups were respectively 16, 14.8 and 11.3 months shorter compared to the equivalent LL group (Figure 4A). The HR and P values are displayed in Figure 4B.
Dp71 expression is an independent prognostic marker of survival in mesothelioma
The characteristics of the high and low DMD gene/transcript expression groups used in the survival analyses are summarised in Figure 5.
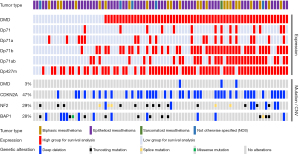
In the mesothelioma cohort analysed here, patients with the non-epithelioid mesothelioma subtype had significantly worse OS compared to those with the epithelioid subtype [HR 1.771; 95% confidence interval (CI): 1.042–3.010; P=0.01; Figure S4], which agreed with the literature (5). However, we did not find any effect for gender, patient age at diagnosis, or tumour stage on the survival of patients in this cohort (P=0.68, P=0.18, and P=0.76, respectively; Figure S4).
Due to the frequent somatic alterations involving the CDKN2A, NF2, and BAP1 genes in mesothelioma and their association with survival (9,10,12,15,16,18,38), we examined whether genomic alterations of these genes have any effect on patient survival in the mesothelioma cohort analysed here.
Deep CDKN2A deletions were associated with significantly worse survival (HR 3.299; 95% CI: 1.952–5.576; P<0.001; Figure S5). Deep NF2 deletions were also associated with worse survival (HR 3.589; 95% CI: 0.9728–13.24; P=0.001; Figure S6A), but we did not find any effects for shallow NF2 deletions or somatic NF2 gene mutations (Figure S6A,S6B). Finally, we did not find any association between the presence of BAP1 CNVs or mutations on mesothelioma survival in this cohort (Figure S7).
Among the analysed DMD gene transcripts, the association between the Dp71 expression and survival remained significant in a multivariate analysis with histology and CDKN2A and NF2 deletion status. Therefore, Dp71 expression is an independent prognostic factor for mesothelioma (Table 1). CDKN2A deletion status also remained significant in this model.
Table 1
| Survival analysis | P value | HR (95% CI) |
|---|---|---|
| Dp71 across all mesothelioma patients | ||
| Dp71 (high vs. low) | 0.008* | 2.291 (1.24, 4.230) |
| CDKN2A deletion status (deep deletions vs. no deep deletions) | <0.001* | 2.966 (1.722, 5.110) |
| Histology (non-epithelioid vs. epithelioid) | 0.06 | 1.599 (0.974, 2.625) |
| NF2 deletion status (deep deletions vs. deep deletions) | 0.15 | 1.832 (0.797, 4.211) |
| Total DMD + Dp71 across all mesothelioma patients | ||
| Total DMD + Dp71 (HH vs. LL) | 0.01* | 3.337 (1.281, 8.689) |
| CDKN2A deletion status (deep deletions vs. no deep deletions) | 0.09 | 2.337 (0.872, 6.269) |
| Histology (non-epithelioid vs. epithelioid) | 0.02* | 2.67 (1.131, 6.300) |
| NF2 deletion status (deep deletions vs. no deep deletions) | 0.73 | 1.446 (0.165, 12.693) |
| Dp71 in mesothelioma patients without CDKN2A deep deletions | ||
| Dp71 (high vs. low) | 0.01* | 3.582 (1.309, 9.802) |
| Histology (non-epithelioid vs. epithelioid) | 0.11 | 1.834 (0.857, 3.926) |
| NF2 deletion status (deep deletions vs. no deep deletions) | 0.19 | 3.949 (0.498, 31.293) |
*, statistically significant P values. DMD, Duchenne muscular dystrophy; HR, hazard ratio; CI, confidence interval; HH, high/high; LL, low/low.
Furthermore, the association between the combined high expression of total DMD gene and Dp71 transcript and survival remained statistically significant in a multivariate analysis (Table 1). Only histology remained significant in this model, while the deletion status of CDKN2A and NF2 did not.
Lastly, the association between the combined high expression of Dp71 and Dp71b transcripts and survival remained statistically significant (HR 2.229; 95% CI: 1.049–4.736; P=0.03; Table S5).
Given our findings that Dp71 (alone or in combination with total DMD or Dp71b) can predict survival independently of deep CDKN2A deletions, we evaluated whether Dp71 can predict survival in mesothelioma patients who do not have deep CDKN2A deletions. We found that Dp71 expression was significantly associated with survival in a multivariate analysis with histology and NF2 deletion status (Table 1). The median OS for the high Dp71 group was 13.61 months compared to 27.16 months for the low Dp71 group, a difference of 13.55 months (Figure S8). Neither histology nor NF2 deletion status remained significant in this model.
Our findings identify the expression levels of Dp71 as a unique biomarker correlating with patient survival in histologically diverse mesotheliomas. Further investigations are needed to understand the potential role for specific dystrophin transcripts in cancer and TME cells and thus in the pathogenesis and/or disease progression of mesothelioma.
Discussion
Key findings
Mesothelioma is a challenging disease presenting difficulties with the assessment of patient prognosis, which has a profound impact on shaping treatment decisions (8). Despite extensive efforts to identify prognostically relevant molecular markers, none are currently used routinely in the clinic.
We found that Dp71 expression is an independent prognostic marker for mesothelioma survival. Median survival of patients with both high Dp71 and high total DMD expression was approximately 2 years shorter than those with low expression. These differences in survival are significant considering the median mesothelioma survival of 7 to 27 months (3). We also found that Dp71 expression is an independent prognostic marker for mesothelioma patients who do not have CDKN2A deep deletions, further emphasising the unique prognostic value offered by Dp71 in this subset of patients. Because no data from corresponding control tissues were available in the TCGA dataset, we were not able to establish whether Dp71 expression is altered in mesothelioma compared to normal pleura.
Concerning the association between Dp71 and mesothelioma, two plausible scenarios warrant consideration. The first scenario suggests that the expression of Dp71 and its distinct splice variants serves as a passive biomarker indicative of mesothelioma status. This expression may alter due to unrelated changes within cancer cells or merely reflect the composition of various TME cell types, which either promote or inhibit tumorigenesis. In the second scenario, a causal mechanism comes into play, wherein the expression of Dp71 isoforms affects cancer—either through a direct impact on cell biology or indirectly by effects in specific TME cells. While the current data does not allow for a conclusive differentiation between these two possibilities, various indicators do exist and provide valuable insights.
Pathways enriched in the DEGs between the high versus low Dp71 expression groups included cell cycle and homologous recombination. These pathways are pivotal in the regulation of cell growth, DNA repair, and genomic stability, processes that are frequently dysregulated in tumours (39,40). Failure of the DNA damage response, comprising a complex network of repair mechanisms and cell cycle checkpoints responsible for detecting and repairing DNA damage, represents one of the important aetiologies for mesothelioma development (41), which highlights the relevance of our findings. The pathways enriched in the DEGs between high versus low Dp71b/ab groups encoding the alternative C-terminus indicated the involvement of this group of variants in the ECM interactions and PI3K-Akt signalling. Indeed, we have found this C-term Dp71 variant to regulate diverse cellular functions including adhesion and proliferation of tumour cells (N. Alnassar, PhD, unpublished data, 2024). Yet another pathway alteration identified is calcium signalling, which is a hallmark abnormality in cells with DMD gene mutations (25). Previously, changes in these pathways have been implicated in the development and progression of cancer (42,43). The PI3K/AKT plays a role in mesothelioma cell viability and survival, and its suppression has been demonstrated to reduce mesothelioma cell viability (41,44). Furthermore, mesothelioma cells exhibit alterations in the expression and/or activation of calcium signalling pathway (41,45). The identification of these pathway alterations in mesothelioma is consistent with our previous findings on pathways altered in different types of primary carcinomas as well as the dystrophic muscle of Duchenne patients (20).
Also, in agreement with our results, Dp71 was previously found to be essential for cell cycle progression (46,47), and its depletion reduced cell proliferation in a range of tumour cells including soft tissue sarcoma (46), myogenic sarcoma (28), lung adenocarcinoma (48) and also PC12 neuronal cells (49). In line with our findings, it was the Dp71 variant (containing exon 78) that was found to play a key role in cell division in PC12 cells (49). Additionally, increased Dp71 expression was associated with poor prognosis of patients with different tumours (19-21).
Regarding the TME cells, we found a positive correlation between Dp71 expression and M2 macrophages and we recently discovered that loss of Dp71 expression impacts the development and key function of mouse macrophages (manuscript in preparation). The presence of high numbers of M2 cells is a negative prognostic factor (50) as these immunosuppressive macrophages produce factors that promote cancer cell survival (37). The expression of Dp71b showed a weak positive correlation with endothelial cells and fibroblasts, while Dp71ab expression correlated with fibroblasts. Fibroblasts contribute to creating an immunosuppressive TME (51) and endothelial cells form neo-vasculature, supplying nutrients to the tumour, as well as facilitating the transit of metastatic cells (52). We also identified a weak negative correlation between total DMD expression and infiltrating NK cells, which control tumour growth and mediate a robust anti-metastatic effect (53). While Dp71 association with multiple TME cells aligns with the ubiquitous expression of this transcript (23), there is currently no strong evidence that fibroblasts and endotheliocytes require this isoform for their proper functioning. On the other hand, given the low prevalence of DMD mutations disrupting Dp71 expression and the short lifespan of Duchenne patients (23), it is conceivable that any potential dysfunctions in these TME cells impacting tumour progression may have gone unnoticed.
Insights into the factors influencing the cellular composition of the TME are becoming increasingly important as they could help identify the responders and non-responders to therapeutic interventions (54). Although at this stage there is no conclusive evidence indicating a specific role of Dp71 in these TME cells, our discovery that the expression of this transcript serves as an independent prognostic biomarker for mesothelioma and may be a revelator of the presence of distinct cellular subsets within the TME, makes it an intriguing target for further research.
Determining the active involvement of Dp71 in mesothelioma and/or TME cell biology, as opposed to its potential role as a passive biomarker reflecting cancer cell characteristics or variations in TME cell numbers, holds the key to gaining valuable insights into the pathobiology of this disease. However, whether it functions or merely serves as an indicator, its significance as a biomarker cannot be understated. Even in its capacity as a straightforward biomarker, Dp71 has the potential to enhance risk stratification and provide crucial guidance for therapeutic strategies.
Strengths and limitations
We present the identification of the Dp71 dystrophin isoform as a unique biomarker allowing the prediction of poor survival across mesotheliomas. The correlation observed between DMD transcript expression and TME components underscores its relevance in investigations aimed at delineating its potential as a predictive marker for mesothelioma immunotherapy. Additionally, as a promising candidate for a novel therapeutic target, the DMD transcript holds considerable potential in advancing the landscape of mesothelioma treatment strategies. We acknowledge that the size of the patient cohort available from the TCGA MESO dataset is relatively small and our findings need to be validated using a larger independent cohort. Furthermore, we were not able to establish whether Dp71 transcript is differentially expressed in mesothelioma compared to normal pleura due to the lack of RNAseq expression data from control tissues in the TCGA dataset.
Comparison with similar researches
Many studies have explored predictive molecular markers for mesothelioma, and while several have been linked to prognosis, none found routine clinical application. These included CDKN2A (9,10) and NF2 (11,12) gene deletions, associated with worse mesothelioma prognosis, and inactivating mutations in the BAP1 gene linked, in some studies, to improved prognosis (16,18). However, these markers have not been demonstrated to outperform the standard clinicopathological parameters in predicting survival. Moreover, the low sensitivity and technical complexity of the detection of these markers restricts their widespread clinical use (14,17).
Explanations of findings
Given the mesothelioma diversity, finding a single biomarker that could inform survival across all types may seem unlikely. However, identification of the DMD gene transcription as such a biomarker not only challenges this view but this association is also explainable at the molecular level. The DMD gene, being the second largest in the human genome and possessing housekeeping characteristics, generates a plethora of functionally distinct isoforms comparable to a versatile Swiss Army knife. With different “business heads” sharing a common handle, its multifunctional housekeeping nature could elucidate its ubiquitous expression. Unsurprisingly, up- and down-regulation of DMD expression levels exhibit an association with survival, regardless of whether they merely mirror the tumour and/or TME status or are directly linked to tumour cell biology. The latter is indicated by the alterations of specific pathways relevant to cancer progression, which were found between the high versus low DMD expression groups. The direct link is further corroborated by functional studies illustrating the dystrophin role in tumours, and that it varies depending on the tumour type (21,22,46,48,49). Additionally, our recent work has extended this this role to specific splice variants (N. Alnassar, PhD, unpublished data, 2024).
Implications and actions needed
To confirm the reliability and clinical relevance of DMD gene transcripts as a prognostic biomarker for mesothelioma, we recognise the necessity for an independent clinical validation study that encompasses a broader spectrum of mesothelioma patients.
Conclusions
Our findings demonstrate that the expression of Dp71 transcript of the DMD gene is an independent prognostic marker for mesothelioma. Thus, quantification of this transcript could potentially be developed into a prognostic and predictive biomarker test to guide treatment decision-making and therapeutic development. The prognostic significance of DMD transcript expression may enable the stratification of patients with either epithelioid or non-epithelioid mesothelioma based on their prognosis. This stratification, in turn, holds the potential to guide therapeutic decisions concerning the intensity of treatment, whether to be initiated upfront or as an option in later palliative phases. Moreover, it bears relevance to determining the optimal intensity of patient follow-up and plays a crucial role in the stratification of patients participating in clinical trials. Further studies unravelling the exact role of dystrophin expression could provide novel insight into the molecular basis of mesothelioma pathology, including its complex tumour environment, and potentially result in the identification of novel therapeutic targets.
Acknowledgments
Funding: The study was supported by University of Portsmouth (under the Strategic PhD studentships scheme to N.A.).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-28/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-28/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-28/coif). N.A. reports University of Portsmouth University Strategic PhD Scholarship. G.L.B. received consulting fees from AstraZeneca, Amgen; fees for speaker bureau or advisory boards from Astellas, Astrazeneca, Amgen and Bayer; fees for travel, accommodations for scientific conferences from Merck and Janssen, outside the submitted work; and had four patents with ST Microelectronics. J.M.J.D. reports consulting fees from DEM Biopharma Inc., Solu Therapeutics, and Keros Therapeutics; and has a 0.25 FTE position with Presage Biosciences. D.C.G. has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang J, Chan SC, Pang WS, et al. Global Incidence, Risk Factors, and Temporal Trends of Mesothelioma: A Population-Based Study. J Thorac Oncol 2023;18:792-802. [Crossref] [PubMed]
- Wadowski B, De Rienzo A, Bueno R. The Molecular Basis of Malignant Pleural Mesothelioma. Thorac Surg Clin 2020;30:383-93. [Crossref] [PubMed]
- Yap TA, Aerts JG, Popat S, et al. Novel insights into mesothelioma biology and implications for therapy. Nat Rev Cancer 2017;17:475-88. [Crossref] [PubMed]
- Janes SM, Alrifai D, Fennell DA. Perspectives on the Treatment of Malignant Pleural Mesothelioma. N Engl J Med 2021;385:1207-18. [Crossref] [PubMed]
- Davidson B. Prognostic factors in malignant pleural mesothelioma. Hum Pathol 2015;46:789-804. [Crossref] [PubMed]
- Blum Y, Meiller C, Quetel L, et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat Commun 2019;10:1333. [Crossref] [PubMed]
- Courtiol P, Maussion C, Moarii M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med 2019;25:1519-25. [Crossref] [PubMed]
- Kirschner M. PL04.04 Prognostic Factors in Malignant Pleural Mesothelioma. J Thorac Oncol 2019;14:S12.
- Dacic S, Kothmaier H, Land S, et al. Prognostic significance of p16/cdkn2a loss in pleural malignant mesotheliomas. Virchows Arch 2008;453:627-35. [Crossref] [PubMed]
- Marshall K, Jackson S, Jones J, et al. Homozygous deletion of CDKN2A in malignant mesothelioma: Diagnostic utility, patient characteristics and survival in a UK mesothelioma centre. Lung Cancer 2020;150:195-200. [Crossref] [PubMed]
- Hiltbrunner S, Fleischmann Z, Sokol ES, et al. Genomic landscape of pleural and peritoneal mesothelioma tumours. Br J Cancer 2022;127:1997-2005. [Crossref] [PubMed]
- Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov 2018;8:1548-65. [Crossref] [PubMed]
- Kinoshita Y, Hamasaki M, Yoshimura M, et al. Hemizygous loss of NF2 detected by fluorescence in situ hybridization is useful for the diagnosis of malignant pleural mesothelioma. Mod Pathol 2020;33:235-44. [Crossref] [PubMed]
- Sa-Ngiamwibool P, Hamasaki M, Kinoshita Y, et al. Usefulness of NF2 hemizygous loss detected by fluorescence in situ hybridization in diagnosing pleural mesothelioma in tissue and cytology material: A multi-institutional study. Lung Cancer 2023;175:27-35. [Crossref] [PubMed]
- Singhi AD, Krasinskas AM, Choudry HA, et al. The prognostic significance of BAP1, NF2, and CDKN2A in malignant peritoneal mesothelioma. Mod Pathol 2016;29:14-24. [Crossref] [PubMed]
- Farzin M, Toon CW, Clarkson A, et al. Loss of expression of BAP1 predicts longer survival in mesothelioma. Pathology 2015;47:302-7. [Crossref] [PubMed]
- Nasu M, Emi M, Pastorino S, et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol 2015;10:565-76. [Crossref] [PubMed]
- Baumann F, Flores E, Napolitano A, et al. Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival. Carcinogenesis 2015;36:76-81. [Crossref] [PubMed]
- Jones L, Naidoo M, Machado LR, et al. The Duchenne muscular dystrophy gene and cancer. Cell Oncol (Dordr) 2021;44:19-32. [Crossref] [PubMed]
- Alnassar N, Borczyk M, Tsagkogeorga G, et al. Downregulation of Dystrophin Expression Occurs across Diverse Tumors, Correlates with the Age of Onset, Staging and Reduced Survival of Patients. Cancers (Basel) 2023;15:1378. [Crossref] [PubMed]
- Naidoo M, Jones L, Conboy B, et al. Duchenne muscular dystrophy gene expression is an independent prognostic marker for IDH mutant low-grade glioma. Sci Rep 2022;12:3200. [Crossref] [PubMed]
- Tan S, Tan J, Tan S, et al. Decreased Dp71 expression is associated with gastric adenocarcinoma prognosis. Oncotarget 2016;7:53702-11. [Crossref] [PubMed]
- Duan D, Goemans N, Takeda S, et al. Duchenne muscular dystrophy. Nat Rev Dis Primers 2021;7:13. [Crossref] [PubMed]
- Austin RC, Howard PL, D'Souza VN, et al. Cloning and characterization of alternatively spliced isoforms of Dp71. Hum Mol Genet 1995;4:1475-83. [Crossref] [PubMed]
- Allen DG, Whitehead NP, Froehner SC. Absence of Dystrophin Disrupts Skeletal Muscle Signaling: Roles of Ca2+, Reactive Oxygen Species, and Nitric Oxide in the Development of Muscular Dystrophy. Physiol Rev 2016;96:253-305. [Crossref] [PubMed]
- Dumont NA, Wang YX, von Maltzahn J, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med 2015;21:1455-63. [Crossref] [PubMed]
- Gosselin MRF, Mournetas V, Borczyk M, et al. Loss of full-length dystrophin expression results in major cell-autonomous abnormalities in proliferating myoblasts. Elife 2022;11:e75521. [Crossref] [PubMed]
- Wang Y, Marino-Enriquez A, Bennett RR, et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet 2014;46:601-6. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 2011;13:36-46. [Crossref] [PubMed]
- Liu P, Sanalkumar R, Bresnick EH, et al. Integrative analysis with ChIP-seq advances the limits of transcript quantification from RNA-seq. Genome Res 2016;26:1124-33. [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Smyth GK, Ritchie M, Thorne N, et al. limma: linear models for microarray and RNA-Seq data user’s guide. Bioinformatics Division, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia; 2002.
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90-7. [Crossref] [PubMed]
- Zaitsev A, Chelushkin M, Dyikanov D, et al. Precise reconstruction of the TME using bulk RNA-seq and a machine learning algorithm trained on artificial transcriptomes. Cancer Cell 2022;40:879-894.e16. [Crossref] [PubMed]
- Hirata E, Sahai E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb Perspect Med 2017;7:a026781. [Crossref] [PubMed]
- Zhang M, Luo JL, Sun Q, et al. Clonal architecture in mesothelioma is prognostic and shapes the tumour microenvironment. Nat Commun 2021;12:1751. [Crossref] [PubMed]
- Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med 2016;6:a026104. [Crossref] [PubMed]
- Stewart MD, Merino Vega D, Arend RC, et al. Homologous Recombination Deficiency: Concepts, Definitions, and Assays. Oncologist 2022;27:167-74. [Crossref] [PubMed]
- Malakoti F, Targhazeh N, Abadifard E, et al. DNA repair and damage pathways in mesothelioma development and therapy. Cancer Cell Int 2022;22:176. [Crossref] [PubMed]
- Noorolyai S, Shajari N, Baghbani E, et al. The relation between PI3K/AKT signalling pathway and cancer. Gene 2019;698:120-8. [Crossref] [PubMed]
- Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, et al. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 2020;11:5120. [Crossref] [PubMed]
- Kanteti R, Riehm JJ, Dhanasingh I, et al. PI3 Kinase Pathway and MET Inhibition is Efficacious in Malignant Pleural Mesothelioma. Sci Rep 2016;6:32992. [Crossref] [PubMed]
- Martinotti S, Patrone M, Moccia F, et al. Targeting Calcium Signalling in Malignant Mesothelioma. Cancers (Basel) 2019;11:1839. [Crossref] [PubMed]
- Mauduit O, Delcroix V, Lesluyes T, et al. Recurrent DMD Deletions Highlight Specific Role of Dp71 Isoform in Soft-Tissue Sarcomas. Cancers (Basel) 2019;11:922. [Crossref] [PubMed]
- Villarreal-Silva M, Suárez-Sánchez R, Rodríguez-Muñoz R, et al. Dystrophin Dp71 is critical for stability of the DAPs in the nucleus of PC12 cells. Neurochem Res 2010;35:366-73. [Crossref] [PubMed]
- Tan S, Tan S, Chen Z, et al. Knocking down Dp71 expression in A549 cells reduces its malignancy in vivo and in vitro. Cancer Invest 2016;34:16-25. [Crossref] [PubMed]
- Villarreal-Silva M, Centeno-Cruz F, Suárez-Sánchez R, et al. Knockdown of dystrophin Dp71 impairs PC12 cells cycle: localization in the spindle and cytokinesis structures implies a role for Dp71 in cell division. PLoS One 2011;6:e23504. [Crossref] [PubMed]
- Ujiie H, Kadota K, Nitadori JI, et al. The tumoral and stromal immune microenvironment in malignant pleural mesothelioma: A comprehensive analysis reveals prognostic immune markers. Oncoimmunology 2015;4:e1009285. [Crossref] [PubMed]
- Mao X, Xu J, Wang W, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 2021;20:131. [Crossref] [PubMed]
- Reymond N, d'Água BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer 2013;13:858-70. [Crossref] [PubMed]
- Melaiu O, Lucarini V, Cifaldi L, et al. Influence of the Tumor Microenvironment on NK Cell Function in Solid Tumors. Front Immunol 2019;10:3038. [Crossref] [PubMed]
- Sadeghi Rad H, Monkman J, Warkiani ME, et al. Understanding the tumor microenvironment for effective immunotherapy. Med Res Rev 2021;41:1474-98. [Crossref] [PubMed]


