
GPRC5A is a potential prognostic biomarker and correlates with immune cell infiltration in non-small cell lung cancer
Highlight box
Key findings
• Low expression of G-protein-coupled receptor family C group 5 type A (GPRC5A) is associated with increased infiltration of immune cells, higher immunophenotype score (IPS), and spatial distribution of programmed cell death-ligand 1 (PD-L1)-positive tumor cells.
What is known and what is new?
• GPRC5A has been linked to the prognoses of some types of cancer.
• This study indicated that low GPRC5A expression is associated with increased immune cell infiltration, including CD8+ T cells, activated CD4+ T cells, and M1 macrophages in non-small cell lung cancer (NSCLC). Moreover, NSCLC tumors with low GPRC5A expression showed an augmented presence of PD-L1-positive cells in the adjacent peripheral regions.
What is the implication, and what should change now?
• GPRC5A is associated with immune cell infiltration, IPS and PD-L1 spatial distribution in NSCLC patients. We speculate that low expression of GPRC5A is associated with immunotherapy, but further validation is still required.
Introduction
Lung cancer is the most common cause of cancer-associated death worldwide, with an estimated 2.2 million new cases and 1.8 million new deaths annually (1). Lung cancer can be classified into two main types, non-small cell lung cancer (NSCLC) (85% of patients) and small cell lung cancer (SCLC) (15%). NSCLC can be further subdivided into lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell carcinoma (2). In more than two decades, the 5-year overall survival (OS) rate for NSCLC patients with metastatic diseases is less than 5% (3). Immune checkpoint inhibitors (ICIs), which can increase the 5-year OS rate for patients with advanced NSCLC, are emerging (4). To date, some biomarkers including programmed cell death-ligand 1 (PD-L1) expression, tumor mutational burden (TMB), and mismatch repair deficiency, have been validated in phase II–III trials of some cancer types and have been widely used (5-8). However, these biomarkers are imperfect because of complex cancer-immune interactions, the tumor microenvironment (TME), and host immunity (9). Hence, continuous research on molecular biomarkers and novel therapies is critical to predicting prognosis and determining personalized treatment.
Immunotherapy is a promising recent approach for treating cancer by activating or boosting the immune system to attack cancer cells (10). Cancer immunotherapy can be administered via the following delivery systems: ICIs, lymphocyte-promoting cytokines, engineered T cells such as chimeric antigen receptor T (CAR-T) and T-cell receptor (TCR) T cells, agonistic antibodies against costimulatory receptors, and cancer vaccines (11). ICIs, such as anti-PD-L1 or anti-programmed cell death 1 (anti-PD-1)/anti-cytotoxic T-lymphocyte-associated antigen-4 (anti-CTLA-4) antibodies, can significantly prolong OS as soon as the 1st line of treatment for advanced NSCLC (12). However, research has shown that most patients with NSCLC develop disease progression during ICIs treatment or after ICIs treatment discontinuation. The Society for Immunotherapy of Cancer (SITC) distinguished three distinct resistance scenarios with anti-PD-1 or PD-L1 treatment: primary resistance, secondary resistance, and progression after treatment discontinuation for any reason (acquired resistance). Primary resistance is defined as evidence of disease progression after receiving at least 6 weeks (two cycles) but not more than 6 months of ICIs treatment. Secondary resistance is defined as disease progression after experiencing clinical benefit [either objective response or stable disease (SD) lasting 6 months or greater]. Acquired resistance is considered in all patients with recurrence after an initial objective response (excluding SD) to ICIs, regardless of the timing of occurrence, and it is a challenging question (13,14). Moreover, searching for a novel epithelial biomarker that can predict immunotherapeutic response is crucial.
G-protein-coupled receptor family C group 5 type A (GPRC5A), a member of the GPCR family also called RAIG1 or RAI3, is a retinoic acid-inducible gene that is downregulated in lung cancer, especially in NSCLC (15). Moreover, GPRC5A knockout mice can spontaneously develop LUAD and the expression of GPRC5A is lower in lung cancer tissue than in normal lung tissue (15). Thus, GPRC5A is considered a tumor suppressor gene in lung cancer (16). Recent studies have shown that GPRC5A has a dual role in different tumor types. The expression of GPRC5A is increased in prostate cancer, ovarian cancer, pancreatic cancer, and gastric cancer (17-20), but decreased in breast cancer (BRCA) and lung cancer (21). According to previous research, the protein product of GPRC5A is significantly associated with adenocarcinoma histology, and GPRC5A expression is negatively correlated with nuclear factor kappa B (NF-κB) expression (22). Activation of NF-κB transcription factors and signaling pathways is associated with innate and adaptive immune responses, indicating that these pathways could be potentially promising immune-related therapeutic targets in lung cancer (23,24). Our results raise new questions regarding the cross-talk between immune cell infiltration and GPRC5A expression in NSCLC and demands more efforts in studies aiming to understand how GPRC5A expression levels are regulated the TME and PD-L1 spatial distribution. We present this article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-739/rc).
Methods
Patients and samples
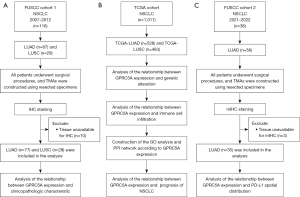
Four tissue microarrays (TMAs) (ten columns and six rows) that contained samples from 87 cases of LUAD and 29 cases of LUSC were included in this study. The TMAs were obtained from Fudan University Shanghai Cancer Center (FUSCC). Each case had two core points to prevent tissue unavailable and information loss. Specimens were collected by surgical resection under a strict standard operating procedure and archived in a formalin-fixed and paraffin-embedded (FFPE) tissue block. Detailed patient information is in Table 1. Finally, a total of 77 LUAD and 29 LUSC samples were included in this analysis. The remaining ten samples were excluded due to tissue unavailable or incomplete patient clinicopathological information. Genomic DNA was extracted from the FFPE tissue samples using the QIAamp DNA Mini Kit (Hilden, Germany), and targeted deep sequencing of mutational hotspots was conducted using a capture-based targeted sequencing panel that included all exons of 68 genes, as described previously (25). For all patients, we collected complete follow-up information, including age, smoking status, pathological stage (pStage), tumor node metastasis (TNM) stage, and epidermal growth factor receptor (EGFR) mutation status. The study included three main NSCLC cohorts (Figure 1), the surgically resected NSCLC cohort (FUSCC cohort 1, n=116) from 2007 to 2012, The Cancer Genome Atlas (TCGA) cohort (n=1,011) and the surgically resected NSCLC cohort (FUSCC cohort 2, n=38) from 2021 to 2022. All specimens were collected from the patients with informed consent, and our study was approved by the Research Ethics Committee of FUSCC (IRB No. 050432-4-1805C). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Characteristics | Total (N=106) | Low expressiona (N=76) | High expression (N=30) | P value |
|---|---|---|---|---|
| Sex | 0.44 | |||
| Male | 83 (78%) | 61 (80%) | 22 (73%) | |
| Female | 23 (22%) | 15 (20%) | 8 (27%) | |
| Age (years) | 0.24 | |||
| <62 | 52 (49%) | 40 (53%) | 12 (40%) | |
| ≥62 | 54 (51%) | 36 (47%) | 18 (60%) | |
| Smoking status | 0.65 | |||
| Never | 32 (30%) | 22 (29%) | 10 (33%) | |
| Ever | 74 (70%) | 54 (71%) | 20 (67%) | |
| Histological type | 0.81 | |||
| ADC | 77 (73%) | 56 (74%) | 21 (70%) | |
| SCC | 29 (27%) | 20 (26%) | 9 (30%) | |
| pStage | 0.047* | |||
| I | 48 (45%) | 31 (40%) | 17 (57%) | |
| II | 20 (19%) | 19 (25%) | 1 (3%) | |
| III | 31 (29%) | 21 (28%) | 10 (33%) | |
| IV | 7 (7%) | 5 (7%) | 2 (7%) | |
| T stage | 0.02* | |||
| T1 | 53 (50%) | 41 (54%) | 12 (40%) | |
| T2 | 36 (34%) | 23 (30%) | 13 (43%) | |
| T3 | 14 (13%) | 12 (16%) | 2 (7%) | |
| T4 | 3 (3%) | 0 (0%) | 3 (10%) | |
| N stage | 0.57 | |||
| N0 | 64 (60%) | 44 (58%) | 20 (67%) | |
| N1 | 9 (9%) | 8 (10%) | 1 (3%) | |
| N2-3 | 33 (31%) | 24 (32%) | 9 (30%) | |
| M stage | >0.99 | |||
| M0 | 97 (92%) | 69 (91%) | 28 (93%) | |
| M1 | 9 (8%) | 7 (9%) | 2 (7%) | |
| Pleural invasion | 0.35 | |||
| Absent | 56 (53%) | 38 (50%) | 18 (60%) | |
| Present | 50 (47%) | 38 (50%) | 12 (40%) | |
| Vascular invasion | 0.53 | |||
| Absent | 73 (69%) | 51 (67%) | 22 (73%) | |
| Present | 33 (31%) | 25 (33%) | 8 (27%) | |
| Perineural invasion | 0.40 | |||
| Absent | 99 (93%) | 72 (95%) | 27 (90%) | |
| Present | 7 (7%) | 4 (5%) | 3 (10%) | |
| EGFR mutation | 0.30 | |||
| Absent | 83 (78%) | 62 (82%) | 21 (70%) | |
| Present | 23 (22%) | 14 (18%) | 9 (30%) |
*, P value <0.05. a, low (H-score <200; IHC negative) or high (≥200; IHC positive) for GPRC5A protein expression. GPRC5A, G-protein-coupled receptor family C group 5 type A; NSCLC, non-small cell lung cancer; FUSCC, Fudan University Shanghai Cancer Center; ADC, adenocarcinoma; SCC, squamous cell carcinoma; pStage, pathological stage; T stage, tumor stage; N stage, node stage; M stage, metastasis stage; EGFR, epidermal growth factor receptor; IHC, immunohistochemistry.
GPRC5A expression in NSCLC in public database
We employed the “Gene_DE” module of Tumor IMmune Estimation Resource 2.0 (TIMER2.0) (http://timer.cistrome.org/) to obtain box-whisker plots of GPRC5A expression between tumor and paracancer tissues in different tumors. Then, we analyzed the GPRC5A protein abundance in Clinical Proteomic Tumor Analysis Consortium (CPTAC) database (https://cprosite.ccr.cancer.gov/).
The correlation between GPRC5A expression and immune cell infiltration
TCGA database (https://portal.gdc.cancer.gov/v1/) was employed to assess the TME score in NSCLC and explore the association between GPRC5A expression, immune cell infiltration, and immune checkpoint expression. Additionally, The Cancer Immunome Atlas (TCIA) database (https://www.tcia.at/home) was utilized to examine the correlation between GPRC5A expression and the immunophenotype score (IPS). Furthermore, we used receiver operating characteristic (ROC) plotter (https://rocplot.com/immune) to predict the relationship between GPRC5A expression and immunotherapy response in pan-cancer. Finally, the TISIDB (http://cis.hku.hk/TISIDB/) database utilized analysis the association of GPRC5A expression and PD-L1 expression and the expression of GPRC5A in different immune subtype of NSCLC.
Gene Ontology (GO) enrichment analysis, genomic alteration analysis and protein-protein interaction (PPI) network of GPRC5A
We conducted GO pathway enrichment analysis to investigate GPRC5A’s potential mechanisms. Initially, genes correlated with GPRC5A were selected via Pearson correlation analysis. Subsequently, we utilized the “clusterProfiler” R package to delve into the functional and molecular mechanisms associated with GPRC5A. Furthermore, gene alteration analysis was executed using the “maftools” package in R software, utilizing RNA seq data from TCGA for LUAD and LUSC. LinkedOmics (https://linkedomics.org/) was used to classify the co-expressed genes of GPRC5A. GPRC5A-associated genes were validated using the Encyclopedia of RNA Interactomes (ENCORI) (https://rnasysu.com/encori/) database. Additionally, we used the cBioPortal for Cancer Genomics (cBioPortal) (https://www.cbioportal.org/) to explore genomic alterations in GPRC5A in NSCLC. To understand the potential interrelationships of GPRC5A with other proteins, we used the STRING database (https://cn.string-db.org/) to establish the PPI network. Finally, Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) was used to analyze the correlation between the expression of GPRC5A and the top five mutated genes.
Survival analysis based on GPRC5A expression in NSCLC
We employed Kaplan-Meier plotter (https://kmplot.com/analysis/), an online platform, to examine gene expression associations with survival in specific tumor types. Utilizing this tool, we assessed the OS of NSCLC patients based on GPRC5A expression. Additionally, we validated OS or disease-free survival (DFS) associations with GPRC5A expression using data from the FUSCC cohort.
Immunohistochemistry (IHC)
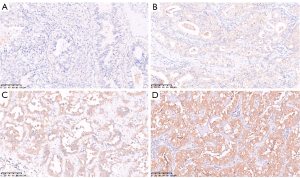
TMA slides were deparaffinized and hydrated, and antigen retrieval was conducted by immersion in sodium citrate buffer (pH 6.0). Then, TMA slides were incubated with an antibody against human GPRC5A (Cell Signaling Technology, Danvers, USA, 12968S, 1:250 dilution) at 4 ℃ overnight. After washing with phosphate buffer saline (PBS) buffer three times, the TMA slides were incubated for 45 minutes with secondary antibody at room temperature (RT). Normal lung tissue was used as a positive control for GPRC5A expression. The intensity of specific staining was characterized as not present (0), weak (1+), distinct (2+), and very strong (3+) based on the average expression in both the cytomembrane and cytoplasm (Figure 2) (26,27). The percentage of cells at different staining intensities was determined by visual assessment, with the score calculated using the formula 1 × (% of 1 + cells) + 2 × (% of 2 + cells) + 3 × (% of 3 + cells). Samples were then classified as either low (H-score <200; IHC negative) or high (H-score ≥200; IHC positive) for GPRC5A protein expression (28).
Multiplex immunohistochemistry (mIHC) and spatial distance analysis
We utilized mIHC to investigate the correlation between GPRC5A expression and PD-L1 expression. Employing an antibody panel consisting of anti-GPRC5A (Cell Signaling Technology, 12968S, 1:250 dilution), anti-PD-L1 (Cell Signaling Technology, 13684S, 1:200 dilution), and anti-pan-CK (Cell Signaling Technology, 4545S, 1:100 dilution), alongside an Opal 7-color manual IHC kit (Akoya, Marlborough, USA, NEL811001KT), we conducted the mIHC following standard procedures as described previously (29). TMA slides were baked for 1 h at 65 ℃ in an oven and deparaffined by xylene and ethanol of gradient concentrations. After applying different primary antibodies (anti-PD-L1, anti-GPRC5A and anti-pan-CK, sequentially), the secondary antibody (Opal Polymer HRP Ms + Rb, Marlborough, USA) was added and incubated, followed by OPAL dye incubation (1:150 dilution). After all antigens being labeled with different antibodies, DAPI (Thermo Fisher, Waltham, USA) was used for nuclei staining. Scanning of the slides was conducted using a Vectra Polaris system, followed by quantitative and spatial analyses facilitated by the HALO software (v3.3.2541.202). Two TMA slides, containing samples from 38 lung cancer cases, were sourced from the FUSCC Tissue Bank. Among these, 35 tumor and adjacent normal tissue samples were eligible for TMA analysis, while three paired samples were excluded due to tissue spot unavailable during technical procedures.
Cell culture
Lung cancer cell lines (A549, H1299, H1975, HCC827 and PC9) and BEAS-2B, a normal human lung epithelial cell line, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in the corresponding medium, RPMI-1640 or Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin (100 U/mL)/streptomycin (100 U/mL) at 37 ℃ in a humidified atmosphere with 5% CO2.
Western blotting
Cells were washed twice with cold PBS and then lysed to extract total proteins using radio immunoprecipitation assay lysis buffer (RIPA buffer) (Thermo Scientific™, Waltham, USA, 89900). These proteins were subjected to western blotting as described before (15). The expression of GPRC5A was measured by a Tanon 5200 system with Chemiluminescent HRP Substrate (Millipore Corporation, Billerica, USA). Antibodies against GPRC5A (sc-390263, 1:800, Santa Cruz Biotechnology, Dallas, Texas, USA) and GAPDH (5174, Cell Signaling Technology, 1:1,000) were used.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
q-PCR was performed with a 7300 Plus Real Time Fluorescence Quantitative PCR Instrument (Applied Biosystems, Carlsbad, USA). The GPRC5A primer sequences (synthesized by Shanghai Tsingke Biotechnology Co., Ltd., Shanghai, China) were as follows: forward primer, 5'-GCCTTCATCATCGGACTGGAC-3', reverse primer, 5'-AGCGATAACATCCTGGACTAGG-3'.
Statistical analysis
Statistical comparisons to determine the correlations between GPRC5A expression and clinicopathologic characteristics were performed using Pearson’s Chi-squared test or Fisher’s exact test according to the situation. The spatial relationship between GPRC5A expression and PD-L1 expression was evaluated using Mann-Whitney test. OS was defined as the date of surgery to the date of death from any cause or the last follow-up. DFS was defined as the date of complete surgical resection to the date of recurrence or death owing to any cause, whichever came first. The Kaplan-Meier method was used to analyze the survival, and the log-rank method was used to examine differences in survival. Statistical analysis was performed using R software (version 4.1.3) or GraphPad Prism (version 8.3.0), and statistical significance was assumed for differences with P<0.05.
Results
Patient characteristics and clinicopathological information
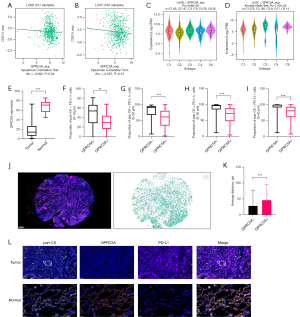
Due to tissue unavailable, a total of 106 patients were analyzed in this study. All patients were recruited from FUSCC during the period spanning 2007 to 2012. The demographic and clinicopathological features, including sex, age, smoking status, histological type, pStage, TNM stage, pleural invasion (PI) status, vascular invasion (VI) status, and perineural invasion (PNI) status, were shown in Table 1. Most patients in this study had LUAD (74%), and more than half of the tumors were in an early stage (65%). Our study showed that the patients with low GPRC5A expression were more likely to be at pStage I or II (P=0.047, Table 1) and T stage I or II (P=0.02, Table 1) in FUSCC cohort. Then, we analyzed the relationship between GPRC5A expression and clinicopathological characteristics in the TCGA cohort (Table S1). The patients with low GPRC5A expression were more likely to be at pStage II (P=0.04, Table S1). In contrast, GPRC5A expression was not significantly associated with sex, age, smoking status, histology type, N stage and M stage in FUSCC cohort or TCGA cohort. Furthermore, we explored the relationship between GPRC5A expression and EGFR mutations in the FUSCC cohort. The results indicated no significant relationship between GPRC5A expression levels and EGFR mutation status (P=0.30, Table 1). Hence, we delved further into the relationship between GPRC5A expression and NSCLC’s treatment.
Pancancer landscape of GPRC5A expression
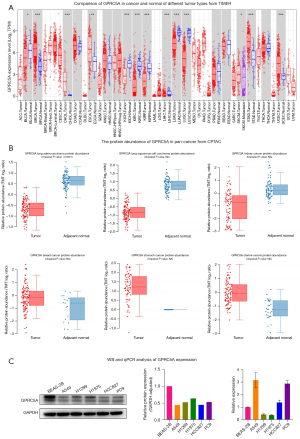
Based on the results from the TIMER database (30), the mRNA expression of GPRC5A showed different levels in 33 types of common cancers. In specific tumor types [BRCA, cholangiocarcinoma (CHOL), esophageal cancer (ESCA), stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC)], GPRC5A expression exhibited significant elevation in cancer tissues compared to adjacent normal tissues. Conversely, in bladder cancer (BLCA), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), LUAD, and LUSC, GPRC5A mRNA levels were notably higher in normal tissues than in cancerous tissues (Figure 3A). Moreover, according to the CPTAC database (31), the GPRC5A protein abundance was increased in cancer tissues compared with normal tissues in BRCA, STAD, and uterine cancer, but was decreased in cancer tissues in LUAD, LUSC, and kidney cancer (Figure 3B), consistent with the previous results. These results showed that GPRC5A may play different roles in different tumors. Then, we found that the expression of GPRC5A in lung cancer cell lines (A549, H1299, H1975, HCC827, PC9) was lower than that in normal lung epithelial cells (BEAS-2B) (Figure 3C) and unedited images of the original western blots in the supplementary material (Figure S1).
Genetic alteration landscape of GPRC5A in NSCLC
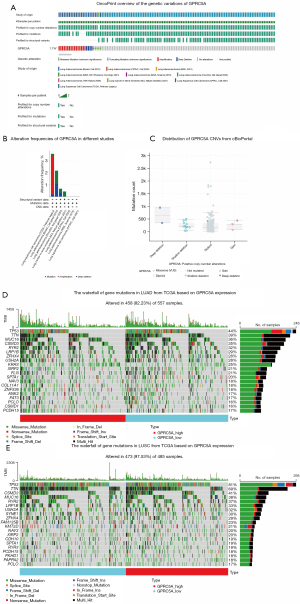
To analyze genetic variations of GPRC5A, we incorporated studies about NSCLC to examine the alterations of GPRC5A expression, covering the Broad, OncoSG, NCI, CPTAC, MSKCC, CPTAC, and TCGA cohorts in cBioPortal (32). The results indicated that the alteration frequency of GPRC5A in NSCLC was 1.7% (Figure 4A) and that mutation, amplification, and deep deletion were common types of alterations, which might be correlated with histological type. The histogram showed that GPRC5A amplification was found mainly in LUSC and that GPRC5A mutation and deep deletion were found mainly in LUAD (Figure 4B). Moreover, there were four types of GPRC5A copy number alterations, namely, gain, diploidy, shallow deletion, and deep deletion, and diploidy was especially prevalent (Figure 4C). To investigate genetic correlations involving GPRC5A alterations, we created waterfall plots using TCGA data in NSCLC (Figure 4D,4E). We selected the top five mutated genes [TP53, titin (TTN), CUB and Sushi multiple domains 3 (CSMD3), mucin16 (MUC16), ryanodine receptor 2 (RYR2)] ranked by mutation for correlation analysis with GPRC5A. The results indicated low correlation coefficients for all genes, with only MUC16 exhibiting a correlation coefficient greater than 0.2, indicative of a weak positive correlation. Further details of the correlation analysis can be found in the supplementary material (Figure S2) (33). Low GPRC5A expression is associated with gene mutations of NSCLC and potentially influence its therapeutic strategies.
The relationship between GPRC5A expression and immune cell infiltration in NSCLC
A comprehensive analysis evaluating GPRC5A expression in NSCLC encompassed assessments of the TME, immune cell infiltration, ICIs, and therapeutic targets using TCGA datasets. The conducted analysis of the TME demonstrated that GPRC5A expression did not affect the matrix score, immune score, or estimate score in TCGA-LUAD (Figure 5A). However, in TCGA-LUSC, we found that low GPRC5A expression could reduce the TME score (Figure 5B). Analysis of immune cell infiltration revealed a negative correlation between GPRC5A expression and the levels of infiltrating plasma cells, CD8+ T cells, activated CD4+ T cells, and M1 macrophages within TCGA-LUAD (Figure 5C). Additionally, in TCGA-LUSC (Figure 5D), a similar negative correlation was observed specifically with the infiltration levels of M1 macrophages. We discovered that decreased expression of GPRC5A may lead to increased infiltration of certain immune cells. Subsequently, we conducted further comparisons to assess the correlation between GPRC5A expression and the levels of infiltration by various immune cell types. Despite observing significant differences, the correlation coefficients between GPRC5A expression and immune cell infiltration levels were low. In TCGA-LUAD, we noted a negative correlation between GPRC5A expression and the levels of activated CD4+ T cells, M1 macrophages, and plasma cells (P=0.01, P<0.001, and P=0.006, respectively), however, the correlation coefficients were low (Figure 5E). Similarly, in TCGA-LUSC, a negative correlation was observed between GPRC5A expression and M1 macrophages (P=0.003), while a positive correlation was found with the infiltration levels of resting mast cells and neutrophils (P<0.001 and P<0.001, respectively) (Figure 5F). However, we identified a correlation coefficient >0.2 only for neutrophils. Additionally, we conducted a comprehensive analysis to investigate the association between GPRC5A expression and TMB in both LUAD and LUSC. We found the coefficients between GPRC5A expression and TMB were low both in LUAD and LUSC (Figure 5G). Furthermore, we conducted an in-depth analysis to explore the relationship between GPRC5A expression and commonly studied ICIs in both LUAD and LUSC. Our findings suggested positive correlations between GPRC5A expression and specific ICIs in LUAD. GPRC5A expression demonstrated significant positive correlations with tumor necrosis factor superfamily 15 (TNFSF15), tumor necrosis factor superfamily 14 (TNFSF14), and tumor necrosis factor superfamily 9 (TNFSF9) (R=0.39, R=0.33, and R=0.33, respectively). Similarly, in LUSC, a positive correlation was observed between GPRC5A expression and TNFSF14 (R=0.36) (Figure 5H). Subsequently, we utilized the TCIA database (34,35) to assess the potential relationship between GPRC5A expression and IPS in NSCLC. Specifically, in LUAD, we observed that low expression of GPRC5A is associated with higher IPS, suggesting that these patients may have responded to anti-CTLA-4 and anti-PD-1 antibodies, but further validation was needed (P=0.03). Conversely, in the case of LUSC, no significant difference in IPS based on GPRC5A expression was observed (P=0.56) (Figure 5I). We assessed the efficacy of GPRC5A expression with anti-PD-1 inhibitors in pan-cancer by ROC plotter (36) and found that low GPRC5A expression is associated with responder, but the area under the curve (AUC) was only 0.588, which may need to be further explored for its predictive value. However, no significant difference was seen for anti-CTLA-4 inhibitors (Figure S3). The comprehensive analysis revealed that in NSCLC, GPRC5A expression exhibited associations with immune cell infiltration and IPS, with specific correlations varying between NSCLC subtypes, highlighting its potential importance as a biomarker and therapeutic target.
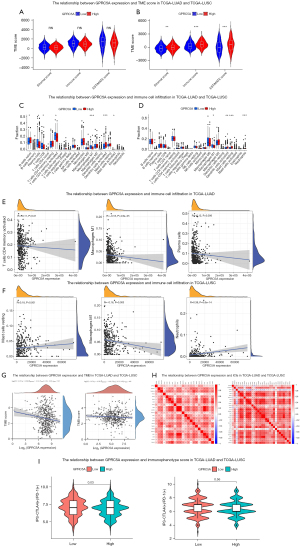
The correlation between GPRC5A and PD-L1 via quantitative and localization
Based on these results, low expression of GPRC5A in lung cancer could increase immune cell infiltration. Combined with the TCGA database, we speculated that there was a certain correlation between the expression of GPRC5A and the response of ICIs, so we further explored the relationship between the expression of GRPC5A and PD-L1. The correlation between GPRC5A and PD-L1 expression was analyzed using the TISDIB database (37). These results indicated little correlation between GPRC5A expression and PD-L1 expression in LUAD (R=−0.092, P=0.04) and LUSC (R=−0.067, P=0.13) (Figure 6A,6B). GPRC5A expression in LUAD and LUSC was analyzed across six immune subtypes (C1-wound healing, C2-IFN-gamma dominant, C3-inflammatory, C4-lymphocyte depleted, C5-immunologically quiet, C6-TGF-β dominant). The findings revealed higher expression of GPRC5A in the C3 and C6 immune subtypes, unaffected by histological type (Figure 6C,6D). To further investigate the impact of GPRC5A expression on PD-L1 expression in localization and its implications on immunotherapeutic response, we conducted mIHC using GPRC5A, PD-L1, pan-CK markers, and DAPI staining on TMAs. Protein expression of GPRC5A was analyzed in 35 pairs of cancer and paracancer tissues, revealing significantly lower expression in tumor areas compared to normal tissue (Figure 6E). Using the spatial analysis module of HALO, we assessed the proportion of pan-CK+PD-L1+ cells within a 40 µm radius of the pan-CK+GPRC5A+ and pan-CK+GPRC5A− cell populations. Remarkably, the pan-CK+GPRC5A− cell population exhibited a higher presence of surrounding pan-CK+PD-L1+ cells within the 0–40 µm range compared to the pan-CK+GPRC5A+ cell population, and this difference was statistically significant (P<0.05) (Figure 6F-6I). By utilizing the Nearest Neighbor Analysis module of HALO software, we examined the spatial association between GPRC5A and PD-L1 expression. The findings revealed that regions exhibiting low GPRC5A expression were in closer proximity to PD-L1-positive regions compared to regions with high GPRC5A expression. This suggested that decreased GPRC5A expression was associated with the spatial distribution of PD-L1-positive regions (Figure 6J,6K) (Figure 6J, 4× magnification). The expression patterns of GPRC5A and PD-L1 in tumor and normal samples are depicted in Figure 6L (20× magnification). Consequently, our findings suggested that reduced GPRC5A expression was associated with the spatial distribution of PD-L1-positive cells, potentially influencing the response of immunotherapy, but further validation is still required.
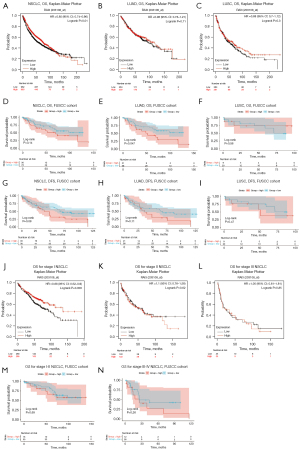
Prognostic evaluation of GPRC5A in NSCLC
To explore the correlation between GPRC5A expression and clinical outcomes in NSCLC patients, we used Kaplan-Meier plotter [including Gene Expression Omnibus (GEO) and TCGA] (38) and FUSCC cohort for prognostic analysis. We examined the OS of individuals diagnosed with LUAD and LUSC using the Kaplan-Meier plotter online tool. Our analysis revealed a significantly poorer OS within the low GPRC5A expression group in the Kaplan-Meier plotter database (P=0.01) (Figure 7A). We conducted additional analysis to assess the prognostic variance between LUAD and LUSC, finding that the disparity was not statistically significant in either LUAD (P=0.71) or LUSC (P=0.30) within the Kaplan-Meier plotter database (Figure 7B,7C). To delve deeper into the prognostic significance of GPRC5A, we utilized TMAs from FUSCC for GPRC5A’s IHC staining, aiming to ascertain its impact on survival rates. Patients were stratified into two categories based on GPRC5A H-scores: those exhibiting high GPRC5A expression (high group, n=30, H-scores ≥200) and those with low GPRC5A expression (low group, n=76, H-scores <200). The findings indicated no notable correlation between GPRC5A expression and OS (Figure 7D, P=0.14) in NSCLC within the FUSCC cohort. Further analysis explored whether the prognosis of NSCLC patients based on GPRC5A expression was influenced by histological type in the FUSCC cohort. The results unveiled a statistically significant disparity in OS concerning LUAD (Figure 7E, P=0.047), whereas the variation in OS for LUSC (Figure 7F, P=0.56) did not exhibit statistical significance within the FUSCC cohort. In addition, we performed DFS analysis on the FUSCC cohort and found that low GPRC5A expression had a better prognosis in both NSCLC and LUAD cohort, rather than LUSC cohort (Figure 7G-7I). Considering that DFS analysis in heterogeneous populations was influenced by various factors, we further conducted univariate Cox and multivariate Cox regression analyses on the FUSCC cohort. The results indicated that low expression of GPRC5A is a protective factor for DFS in NSCLC, but further validation is still required (Table S2). Given the correlation between GPRC5A expression and the pStage of NSCLC observed in both the TCGA and FUSCC cohorts, we conducted a prognostic analysis based on pStage in both the Kaplan-Meier plotter database and the FUSCC cohort. Our findings unveiled a stage-dependent variation in the prognostic influence of GPRC5A expression. In patients with pStage I NSCLC in the Kaplan-Meier plotter, low GPRC5A expression is correlated with decreased OS time (Figure 7J, P=0.006). However, for pStage II–III NSCLC in Kaplan-Meier plotter, this difference was not statistically significant (Figure 7K,7L). Interestingly, within the FUSCC cohort, there was no significant discrepancy in OS based on pStage (Figure 7M,7N). These inconsistencies suggest that the prognostic impact of GPRC5A expression in NSCLC is not uniform across different stages. Further exploration is warranted to elucidate its underlying mechanisms.
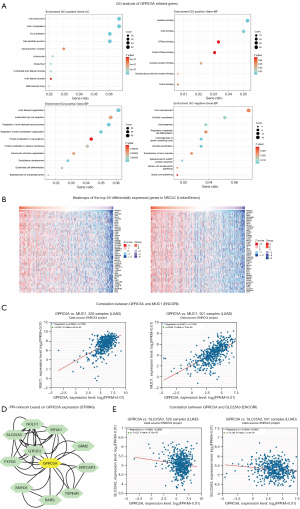
GO analysis and PPI network construction about GPRC5A
To investigate GPRC5A’s potential functions and molecular mechanisms in NSCLC, we initially conducted Pearson correlation analysis to identify genes associated with GPRC5A (top 500 in LUAD and top 500 in LUSC). Subsequently, we performed GO enrichment analysis on the identified genes. The enriched GO terms included cell-cell junction, focal adhesion, GTPase binding, actin filament organization, regulation of actin filament-based process, and axon development (Figure 8A). Additionally, we delved into GPRC5A expression in NSCLC using data mining in the TCGA database’s NSCLC cohort through LinkedOmics (39). The generated heatmap revealed a notable correlation coefficient between mucin 1 (MUC1) and GPRC5A expression (Figure 8B), a finding corroborated by validation in ENCORI (40) (Figure 8C). We established a PPI network, identifying ten genes associated with GPRC5A protein function (Figure 8D). Our data revealed significant correlations between GPRC5A and the following proteins: metabotropic glutamate receptor 2 (GRM2), eukaryotic initiation factor 4A-I (EIF4A1), general transcription factor IIF subunit 2 (GTF2F2), SET and MYND domain-containing protein 5 (SMYD5), FXYD domain-containing ion transport regulator 3 (FXYD3), phosphate carrier protein, mitochondrial (SLC25A3), arginine-tRNA ligase (RARS), keratinocyte associated protein 3 (KRTCAP3), thrombospondin-type laminin G domain and EAR repeat-containing protein (TSPEAR), and nucleolar and coiled-body phosphoprotein 1 (NOLC1). These correlations were notably high, with correlation coefficients ranging from 0.609 to 0.819, respectively. Then, we performed correlation analysis of these key genes (Figure S4) and found that SLC25A3 expression was negatively correlated with GPRC5A expression (Figure 8E). These findings provide additional evidence supporting the association between GPRC5A expression and the effectiveness of immunotherapy in NSCLC.
Discussion
GPRC5A acts as a tumor suppressor in the development of NSCLC (41), and it is downregulated in NSCLC tissue and lung cancer cell lines (15). Our results showed that deletion of GPRC5A might be one of the main mechanisms underlying the dysfunction of GPRC5A in NSCLC. Previous studies confirmed that GPRC5A deletion can enhance the transformed phenotype in normal and malignant lung epithelial cells through the Stat3 signaling pathway (42). GPRC5A deficiency with tobacco carcinogen exposure results in the development of LUAD tumors with somatic mutations in Kirsten rat sarcoma viral oncogene (KRAS), in which the cells exhibit some characteristics of cancer stem cells (CSCs) to increase tumorigenesis in a syngeneic setting in vivo (43). In this study, we investigated the expression of GPRC5A in NSCLC and evaluated the relationship of GPRC5A expression and immune cell infiltration, IPS and PD-L1 spatial distribution. We analyzed the expression and prognostic value of GPRC5A in NSCLC by using FUSCC cohort and public databases. Our results showed that NSCLC with low GPRC5A expression was prone to earlier pStage, and correlated with increased immune cell infiltration, higher IPS and PD-L1 spatial distribution in NSCLC.
In our study, we used TIMER database and CPTAC to analyze the expression profiles of GPRC5A in various human tumors. The results showed GPRC5A gene expression was lower in BLCA, KICH, KIRC, KIRP, LUAD, and LUSC than normal tissues. To further validate the expression of GPRC5A in NSCLC, we conducted qPCR and western blot. Our results showed GPRC5A had a high expression in BEAS-2B. All these results suggested that GPRC5A might act as a promising biomarker, but further validation is still required.
Immunotherapy is an emerging therapeutic strategy for patients with advanced NSCLC (44). Previous studies have shown that high TMB and PD-L1 expression are predictive of a benefit from ICIs treatment in NSCLC (45). It has been shown that PD-L1 expression and TMB can predict immune efficacy, but these markers are not good enough to predict immune efficacy, and some patients develop immunotherapeutic resistance within a short period (45,46). Currently, nearly all patients with metastatic NSCLC who do not harbor targetable oncogenes receive PD-1 or PD-L1 therapy in the first-line setting (47).
To further study the underlying mechanisms and relationships of GPRC5A expression in NSCLC TME and PD-L1 expression, some immune-related analyses were performed based on public databases. At the same time, mIHC was performed based on the FUSCC cohort to explore the relationship between GPRC5A expression and PD-L1 expression. Firstly, in our analysis of immune cell infiltration in NSCLC, we observed heightened presence of CD8+ T cells, activated CD4+ T cells, and M1 macrophages in patients exhibiting low GPRC5A expression. Studies indicated that heightened infiltration of CD8+ T cells is linked to improved immunotherapeutic efficacy (48), CD4+ T cells have demonstrated their capacity to promote tumor rejection in an IFN-γ-dependent manner by targeting the tumor stroma and suppressing angiogenesis (49). Additionally, an elevated presence of M1-like macrophages within NSCLC tumors appears to associate with improved patient outcomes and increased responsiveness to immunotherapy (50). Secondly, our investigation revealed an association between increased IPS and lower levels of GPRC5A expression. Previous studies showed that GPRC5A may be related to tumor immunity and CSC expansion (51). Additionally, bioinformatics analysis (Figure 6A,6B) reveals a weak coefficient between GPRC5A expression and PD-L1 expression in both LUAD and LUSC. It is hard to draw a solid conclusion just based on a coefficient. The TCGA database only distinguish the LUAD and LUSC cohort, some cases lacking the detail mutant background, which is hard for us to further discuss in different subgroups. The correlation coefficient may be tightly related to different subgroups. We will follow this direction to study in the future. Finally, our mIHC analysis revealed significantly lower GPRC5A expression in tumor areas compared to normal tissue. Remarkably, the spatial analysis demonstrated that regions with low GPRC5A expression exhibited a higher presence of PD-L1-positive cells in proximity. This indicates a potential spatial relationship between decreased GPRC5A expression and the distribution of PD-L1-positive cells within the TME. The altered spatial distribution of PD-L1-positive cells in the context of reduced GPRC5A expression might be associated with immunotherapy, but further validation is still required. These findings open avenues for exploring GPRC5A as a potential biomarker and therapeutic target in the context of immunotherapy for NSCLC patients, but further validation was needed.
While our study sheds light on the potential impact of GPRC5A expression on immune cell infiltration in NSCLC, it is essential to acknowledge certain limitations that warrant consideration. Firstly, our analysis predominantly relies on retrospective data and bioinformatics approaches, which may introduce inherent biases or limitations associated with data quality, sample sizes, and variations in methodologies across different datasets. Secondly, while bioinformatics analyses provide valuable insights into immune cell infiltration and molecular correlations, further experimental validation, such as in vitro and in vivo studies, are necessary to confirm the functional relevance of GPRC5A in the TME and its direct influence on immunotherapeutic responses. Moreover, our study primarily focused on associations and correlations without delineating causative relationships, necessitating additional mechanistic investigations to comprehensively understand the precise role of GPRC5A in modulating immune responses and its impact on immunotherapy efficacy in NSCLC. Furthermore, our findings primarily highlight correlations between GPRC5A expression and immune-related parameters; however, clinical response to immunotherapy is multifactorial and influenced by various other factors not solely dependent on GPRC5A expression. Next than, the spatial mIHC analysis, while informative, provides associations but it does not establish a direct causal relationship between GPRC5A expression and alterations in the spatial distribution of PD-L1-positive regions. Finally, the main limitation of this study is the lack of information on immunotherapy within FUSCC cohort, which makes it challenging to draw direct conclusions regarding the association between GPRC5A expression and immune therapy. However, by exploring from pan-cancer data to predict the relationship between GPRC5A expression and immunotherapy response, we speculate that low expression of GPRC5A may be associated with anti-PD-1 response, which warrants further evaluations in an immunotherapy cohort. Addressing these limitations through further comprehensive and prospective studies will provide more definitive insights into the role of GPRC5A in NSCLC immunotherapy and its potential as a predictive biomarker or therapeutic target.
Conclusions
In summary, we observed that low expression of GPRC5A is associated with increased infiltration of immune cells, higher IPS, and spatial distribution of PD-L1-positive tumor cells in NSCLC. These findings suggest the potential utility of GPRC5A as a predictive marker or therapeutic target in NSCLC, complementing established markers like PD-L1 and TMB. Future prospective studies are essential to validate these findings, providing crucial insights to refine immunotherapy strategies for optimizing NSCLC treatment responses.
Acknowledgments
The authors express gratitude to FUSCC Tissue Bank.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-739/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-739/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-739/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-739/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All specimens were collected from the patients with informed consent, and our study was approved by the Research Ethics Committee of FUSCC (IRB No. 050432-4-1805C). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. [Crossref] [PubMed]
- Skribek M, Rounis K, Tsakonas G, et al. Complications following novel therapies for non-small cell lung cancer. J Intern Med 2022;291:732-54. [Crossref] [PubMed]
- Garon EB, Hellmann MD, Rizvi NA, et al. Five-Year Overall Survival for Patients With Advanced Non-Small-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J Clin Oncol 2019;37:2518-27. [Crossref] [PubMed]
- Felip E, Altorki N, Zhou C, et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet 2021;398:1344-57. [Crossref] [PubMed]
- Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353-65. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Andre T, Amonkar M, Norquist JM, et al. Health-related quality of life in patients with microsatellite instability-high or mismatch repair deficient metastatic colorectal cancer treated with first-line pembrolizumab versus chemotherapy (KEYNOTE-177): an open-label, randomised, phase 3 trial. Lancet Oncol 2021;22:665-77. [Crossref] [PubMed]
- Zhou F, Qiao M, Zhou C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell Mol Immunol 2021;18:279-93. [Crossref] [PubMed]
- Kennedy LB, Salama AKS. A review of cancer immunotherapy toxicity. CA Cancer J Clin 2020;70:86-104. [Crossref] [PubMed]
- Riley RS, June CH, Langer R, et al. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov 2019;18:175-96. [Crossref] [PubMed]
- Lee CK, Man J, Lord S, et al. Clinical and Molecular Characteristics Associated With Survival Among Patients Treated With Checkpoint Inhibitors for Advanced Non-Small Cell Lung Carcinoma: A Systematic Review and Meta-analysis. JAMA Oncol 2018;4:210-6. [Crossref] [PubMed]
- Passaro A, Brahmer J, Antonia S, et al. Managing Resistance to Immune Checkpoint Inhibitors in Lung Cancer: Treatment and Novel Strategies. J Clin Oncol 2022;40:598-610. [Crossref] [PubMed]
- Schoenfeld AJ, Antonia SJ, Awad MM, et al. Clinical definition of acquired resistance to immunotherapy in patients with metastatic non-small-cell lung cancer. Ann Oncol 2021;32:1597-607. [Crossref] [PubMed]
- Tao Q, Fujimoto J, Men T, et al. Identification of the retinoic acid-inducible Gprc5a as a new lung tumor suppressor gene. J Natl Cancer Inst 2007;99:1668-82. [Crossref] [PubMed]
- Wang J, Farris AB, Xu K, et al. GPRC5A suppresses protein synthesis at the endoplasmic reticulum to prevent radiation-induced lung tumorigenesis. Nat Commun 2016;7:11795. [Crossref] [PubMed]
- Sawada Y, Kikugawa T, Iio H, et al. GPRC5A facilitates cell proliferation through cell cycle regulation and correlates with bone metastasis in prostate cancer. Int J Cancer 2020;146:1369-82. [Crossref] [PubMed]
- Moyano-Galceran L, Pietilä EA, Turunen SP, et al. Adaptive RSK-EphA2-GPRC5A signaling switch triggers chemotherapy resistance in ovarian cancer. EMBO Mol Med 2020;12:e11177. [Crossref] [PubMed]
- Duggan SN, Ewald N, Kelleher L, et al. The nutritional management of type 3c (pancreatogenic) diabetes in chronic pancreatitis. Eur J Clin Nutr 2017;71:3-8. [Crossref] [PubMed]
- Liu H, Zhang Y, Hao X, et al. GPRC5A overexpression predicted advanced biological behaviors and poor prognosis in patients with gastric cancer. Tumour Biol 2016;37:503-10. [Crossref] [PubMed]
- Yang L, Ma T, Zhang J. GPRC5A exerts its tumor-suppressive effects in breast cancer cells by inhibiting EGFR and its downstream pathway. Oncol Rep 2016;36:2983-90. [Crossref] [PubMed]
- Fujimoto J, Kadara H, Garcia MM, et al. G-protein coupled receptor family C, group 5, member A (GPRC5A) expression is decreased in the adjacent field and normal bronchial epithelia of patients with chronic obstructive pulmonary disease and non-small-cell lung cancer. J Thorac Oncol 2012;7:1747-54. [Crossref] [PubMed]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 2006;441:431-6. [Crossref] [PubMed]
- Yu H, Lin L, Zhang Z, et al. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther 2020;5:209. [Crossref] [PubMed]
- Wang Y, Xue Q, Zheng Q, et al. SMAD4 mutation correlates with poor prognosis in non-small cell lung cancer. Lab Invest 2021;101:463-76. [Crossref] [PubMed]
- Cheng Y, Lotan R. Molecular cloning and characterization of a novel retinoic acid-inducible gene that encodes a putative G protein-coupled receptor. J Biol Chem 1998;273:35008-15. [Crossref] [PubMed]
- Lin X, Zhong S, Ye X, et al. EGFR phosphorylates and inhibits lung tumor suppressor GPRC5A in lung cancer. Mol Cancer 2014;13:233. [Crossref] [PubMed]
- Mazières J, Brugger W, Cappuzzo F, et al. Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: re-analysis of the SATURN study. Lung Cancer 2013;82:231-7. [Crossref] [PubMed]
- Sun H, Wang X, Zhang X, et al. Multiplexed immunofluorescence analysis of CAF-markers, EZH2 and FOXM1 in gastric tissue: associations with clinicopathological parameters and clinical outcomes. BMC Cancer 2022;22:1188. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Thangudu RR, Rudnick PA, Holck M, et al. Proteomic Data Commons: A resource for proteogenomic analysis. Cancer Res 2020;80:Abstract nr LB-242.
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016;165:35-44. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Kovács SA, Fekete JT, Győrffy B. Predictive biomarkers of immunotherapy response with pharmacological applications in solid tumors. Acta Pharmacol Sin 2023;44:1879-89. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Győrffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharmacol 2024;181:362-74. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Jin E, Wang W, Fang M, et al. Lung cancer suppressor gene GPRC5A mediates p53 activity in non small cell lung cancer cells in vitro. Mol Med Rep 2017;16:6382-8. [Crossref] [PubMed]
- Chen Y, Deng J, Fujimoto J, et al. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent Stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res 2010;70:8917-26. [Crossref] [PubMed]
- Daouk R, Hassane M, Bahmad HF, et al. Genome-Wide and Phenotypic Evaluation of Stem Cell Progenitors Derived From Gprc5a-Deficient Murine Lung Adenocarcinoma With Somatic Kras Mutations. Front Oncol 2019;9:207. [Crossref] [PubMed]
- Proto C, Ferrara R, Signorelli D, et al. Choosing wisely first line immunotherapy in non-small cell lung cancer (NSCLC): what to add and what to leave out. Cancer Treat Rev 2019;75:39-51. [Crossref] [PubMed]
- Negrao MV, Skoulidis F, Montesion M, et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J Immunother Cancer 2021;9:e002891. [Crossref] [PubMed]
- Huang MY, Jiang XM, Wang BL, et al. Combination therapy with PD-1/PD-L1 blockade in non-small cell lung cancer: strategies and mechanisms. Pharmacol Ther 2021;219:107694. [Crossref] [PubMed]
- Peters S, Reck M, Smit EF, et al. How to make the best use of immunotherapy as first-line treatment of advanced/metastatic non-small-cell lung cancer. Ann Oncol 2019;30:884-96. [Crossref] [PubMed]
- Hurkmans DP, Kuipers ME, Smit J, et al. Tumor mutational load, CD8(+) T cells, expression of PD-L1 and HLA class I to guide immunotherapy decisions in NSCLC patients. Cancer Immunol Immunother 2020;69:771-7. [Crossref] [PubMed]
- Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res 2014;2:91-8. [Crossref] [PubMed]
- Liu J, Geng X, Hou J, et al. New insights into M1/M2 macrophages: key modulators in cancer progression. Cancer Cell Int 2021;21:389. [Crossref] [PubMed]
- Guo W, Hu M, Wu J, et al. Gprc5a depletion enhances the risk of smoking-induced lung tumorigenesis and mortality. Biomed Pharmacother 2019;114:108791. [Crossref] [PubMed]


