
Predictive value of delta-radiomic features for prognosis of advanced non-small cell lung cancer patients undergoing immune checkpoint inhibitor therapy
Highlight box
Key findings
• We developed a delta-radiomics model with good performance in predicting outcomes in advanced non-small-cell lung carcinoma (NSCLC) patients undergoing immune checkpoint inhibitors (ICIs) therapy, which was superior to the clinical model and pre-treatment radiomics model, respectively.
What is known and what is new?
• Radiomics was applied in the prediction of NSCLC survival before.
• This study applied delta-radiomics features for survival prediction in advanced NSCLC patients, and achieve satisfactory predictive efficiency.
What is the implication, and what should change now?
• As an alternative method for predicting response to ICI is urgently required, optimal delta-radiomic features could improve clinical decision-making to continue systemic therapies and forecast overall survival.
Introduction
Immune checkpoint inhibitors (ICIs), including the programmed cell death protein 1 (PD-1) or its ligand (PD-L1), have markedly attracted oncologists’ attention in the treatment of diverse types of cancer (1,2), especially in the therapy of advanced non-small-cell lung carcinoma (NSCLC) (3). To date, the response rates to the ICI in patients with NSCLC have still remained unsatisfactory (10–40%), depending on each patient’s clinical condition (4). In addition, as the expression levels of biomarkers, such as PD-L1 expression, cannot accurately predict the effectiveness of immunotherapy in advanced NSCLC patients, selection of the most appropriate therapy for such patients remains a clinical challenge.
The immune Response Evaluation Criteria in Solid Tumors (iRECIST) represents an enhanced and revised iteration of the RECIST guidelines. Its widespread adoption in clinical trials worldwide is owed to its improved and standardized assessment of tumor response, specifically incorporating immune-based treatment responses like hyperprogression and pseudoprogression (5,6). Nonetheless, the iRECIST criteria overlook changes in various tumor characteristics beyond size, such as tumor viability, metabolic activity, and tumor density, which could be pertinent to tumor response (7,8). Hence, there is an urgent need for an alternative approach to anticipate response to ICI therapy.
ICIRadiomics involves the extraction of numerous features from medical images through a set of techniques (9). Numerous studies have highlighted the promising potential of radiomics in predicting gene mutations (10,11), lymph node metastasis (12), therapeutic response (13,14), and clinical prognosis (15) of patients with lung cancer. Recently, delta-radiomics features have markedly attracted clinicians’ attention, which show changes in the radiomics features between baseline and follow-up data during treatment. These features have proven effective in enhancing the performance of diagnostic models for differentiating ground-glass nodules (GGNs) (16) and predicting treatment response (17). However, to date, few studies have predicted response to ICI therapy and overall survival (OS) of patients with NSCLC (8). Moreover, one of the main limitations of previous studies was the lack of an external dataset for model validation, highly restricting the generalizability of their proposed radiomics models.
The present study aimed to explore the role of delta-radiomics features in predicting the clinical outcomes of patients with advanced NSCLC who received ICI therapy. We present this article in accordance with the TRIPOD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-7/rc).
Methods
Approval for this retrospective study was obtained from the Ethics Committee of Wuhan Union Hospital (Wuhan, China) (Approval No. S0516), with the requirement for written informed consent waived. Wuhan Tongji Hospital was also informed and agreed the study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Patients and inclusion criteria
Data of a total of 1,207 patients with advanced NSCLC (IIIB–IV stages) from two medical institutions (Wuhan Union Hospital, 836; Wuhan Tongji Hospital, 371) who were confirmed by needle biopsy or bronchofibroscopy from September 2018 to July 2020 and from September 2018 to September 2019 were retrospectively analyzed. As showed in Figure 1, the inclusion criteria were as follows: (I) patients who received at least two cycles of ICI therapy; (II) patients with available computed tomography (CT) findings before immunotherapy; (III) patients with available follow-up data; and (IV) patients with measurable tumors. Thus, in total, 66 patients were excluded (45 of 178 patients from Wuhan Union Hospital and 21 of 67 patients from Wuhan Tongji Hospital). The exclusion criteria are summarized in Figure 1. A total of 133 patients from Wuhan Union Hospital (Database 1) and 46 patients from Wuhan Tongji Hospital (Database 2) were eventually included. Patients in the Database 1 group were randomly assigned into training database (106 patients) and validation dataset (27 patients), with a ratio of 8:2. Meanwhile, patients in the Database 2 group were allocated into testing dataset.

The patients’ clinical characteristics, pathological findings, and therapeutic regimen, including age, gender, smoking history, cancer type, tumor-node-metastasis (TNM) staging, PD-1 expression level, and data related to dosage and type of drug were recorded. A history of smoking was defined as lifetime exposure to more than 100 cigarettes. The eighth edition of TNM stage system for NSCLC presented by the International Association for the Study of Lung Cancer was adopted (18).
Measurement endpoints and follow-up time
The primary endpoint, progression-free survival (PFS), was defined as the duration from the initiation of treatment to disease progression or death, whichever occurred first. Subjects who remained free of progression and were alive at their last follow-up were treated as censored observations, with the time to their last follow-up utilized for analysis. According to the iRECIST guidelines (5,6), patients who had progressive disease (PD) were regarded as “non-responders”, accompanying with a 20% increase in the sum of the longest diameter of target lesions (an absolute increase of 5 mm), and patients with complete response, partial response or stable disease were considered as “responders” (Figure 2). There were two special cases: (I) ICI therapy was performed again after the first evaluation of PD to consider the possible clinical benefits; if the secondary efficacy evaluation was still PD, the real-time progression time of PFS was calculated based on the time of first disease progression. (II) After the first evaluation of PD, ICI treatment was carried out again considering the possible clinical benefits, and the second evaluation was non-PD pseudo progress. The secondary endpoint was OS, which was measured as the time from the start of treatment to death. Subjects who were alive as of their last follow-up were considered censored, and time to the last follow-up was used. Follow-up time was measured from the initiation of immunotherapy until the last follow-up or death.
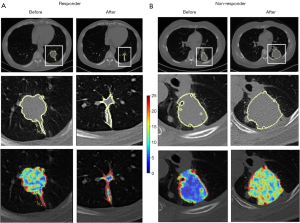
CT scan
All participants underwent CT scan at baseline and immediately after 2–3 cycles (6–8 weeks) of ICI treatment. All images were obtained on one of the four multi-slice spiral CT scanners (SOMATOM Definition AS+, Siemens Healthineers, Erlangen, Germany; uCT 780, United Imaging, Shanghai, China; Optima 660, GE, Tokyo, Japan) at our institutions, using the following parameters: detector collimation width, 64 mm × 0.6 mm and 128 mm × 0.6 mm; tube voltage, 120 kV. The tube current was regulated by an automatic exposure control system (CARE Dose 4D; Siemens Healthcare, Erlangen, Germany). Images were reconstructed at a slice thickness of 1.5 or 1 mm with an increment of 1.5 or 1 mm. No contrast medium was used. Then, all images were resampled with a slice thickness of 1.5 mm with the same increment to ensure uniform voxel size (19). The three-dimensional (3D)-slicer software was utilized to import Digital Imaging and Communications in Medicine (DICOM) images from the picture archiving and communication system (PACS).
Radiomics feature extraction
Three junior radiologists (X.H., Y.Z., and X.J., with 5, 3, and 2 years of experience in thoracic imaging, respectively) semi-automatically delineated the regions of interest (ROIs) on CT images, layer-by-layer using 3D-slicer software. Then, the volumes of interest (VOIs) of tumors were automatically reconstructed using the 3D-slicer software (Figure 3).

Two senior radiologists (H.S. and Y.W., with 32 and 13 years of experience in thoracic imaging, respectively) were tasked with verifying the segmentation of tumors. Any discrepancies were addressed through additional corrections. All above radiologists were blinded to clinical and outcome findings.
Radiomics feature extraction was conducted using the Philips Radiomics Tool (Philips Healthcare, Shanghai, China), with core feature calculations relying on a third-party Python library known as pyRadiomics (20). A total of 1,037 radiomic features, encompassing direct, wavelet-transformed, logarithmic-transformed, and gradient-filtered features, were extracted for each VOI (refer to Figure 3, with additional details available at https://pyradiomics.readthedocs.io/en/latest/features.html). These 1,037 radiomic features could be categorized based on image type: 93 gradient features, 93 LoG features, 107 original features, and 744 wavelet features. Regarding feature class, there were 198 first-order features, 264 gray-level co-occurrence matrix (GLCM) features, 154 gray-level dependence matrix (GLDM) features, 176 gray-level run length matrix (GLRLM) features, 176 gray-level size zone matrix (GLSZM) features, 55 neighboring gray-tone difference matrix (NGTDM) features, and 14 shape features. All features were extracted from the 3D volume. In addition, intra-observer and interobserver variabilities were analyzed in 30 randomly selected CT images (Database 1 and Database 2 both equally included 15 cases). One observer repeated the segmentation after 1 week for intra-observer variability analysis, while the other observer performed the segmentation again using the same method. An intraclass correlation coefficient (ICC) cutoff value of >0.8 was utilized to identify stable and reproducible features.
Delta-radiomics features
We obtained stable and reproducible features after ICI therapy.
For patients who underwent CT scans at baseline and follow-up, radiomics features were extracted from these two time points, respectively. The delta-radiomics features were defined as the relative net change of radiomics features between the two time points (17):
Statistical analysis
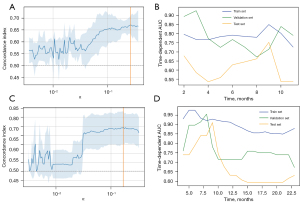
The SPSS software (SPSS 21.0 for Windows, IBM, Chicago, IL, USA), as well as “scikit-survival” (21), packages in Python were used to perform the statistical analysis. Heatmap of delta_wavelet_LLL_glcm_contrast features in the pre- and post-treatment CT scans were illustrated using the “scikit-image” package (22). Continuous variables were presented as median [interquartile range (IQR)], while categorical variables were expressed as frequency (percentage). The Student’s t-test and Mann-Whitney U test was applied to compare differences between the two groups, and the Fisher exact test was used to compare categorical variables. All radiomics features were normalized to the z-score. Pearson’s correlation analysis was used to eliminate the radiomics features with high correlation (r>0.8), thereby excluding redundant variables to z score (23). And inter-observer variability analysis were employed to exclude features of low reliability (ICC <0.8) (24). When two features were highly correlated, the feature that showed stronger correlation with PSF and OS was chosen. The least absolute shrinkage and selection operator (LASSO) was employed to identify features for subsequent assessment (25). Five-fold cross-validation and maximum Harrell’s concordance index (C-index) were used as the feature filtering criteria. The predictive model was created using a Cox proportional hazards (PHs) regression. In addition, three models were established on each iteration using only the high-risk clinical characteristics, pretreatment CT radiomics, and delta-radiomics covariates. Four models for independent prediction of PFS and OS were constructed based on three subsets of features: a clinical model based on high-risk characteristics, a radiomics model based on pretreatment CT radiomics features, and a delta-radiomics model based on the most influential delta-radiomics features. Then, we incorporated the selected radiomics features and clinical features into the LASSO-Cox risk regression analysis, thus obtaining a mix model, as previously reported (26). Moreover, C-index and time-independent receiver operating characteristic (ROC) curve analysis were employed to assess the discriminatory ability of the three models. Patients were stratified as high- or low-risk of poor PFS and OS based on their rank at above or below the median prediction for each model. The Kaplan-Meier curve was plotted for data stratification, and the log-rank test was utilized to determine whether stratified data were significant (P<0.05).
Results
Participants’ characteristics at baseline
A total of 179 eligible patients [median age, 63 (IQR: 56, 67) years old; male (n=149) vs. female (n=30)] were involved. As listed in Table 1, the main pathological type of tumors was adenocarcinoma (108/179, 60.3%), while more than half of patients (116/179, 64.8%) had stage IV NSCLC. Among 68 patients with known PD-1 expression, positive PD-1 expression was dominant (51/68, 75.0%). For PFS, disease progression was found in 50 of 179 (27.9%) cases at follow-up CT scans. The median PFS (after therapy) was 7.0 (IQR: 3.4, 9.1) (range, 1.4–13.2) months. Regarding therapeutic strategy, 66 (36.9%) patients received ICI monotherapy, and 113 (63.1%) patients were treated with combination of immunotherapy and chemotherapy. More specifically, 161 (89.9%) patients received anti-PD-1 agents (e.g., nivolumab, pembrolizumab, camrelizumab, tislelizumab, toripalimab, etc.), and 18 (10.1%) patients received anti-PD-L1 agents (e.g., atezolizumab and durvalumab). Regarding OS, 91 of 179 (50.8%) patients died during follow-up. The median survival time was 12 (IQR: 8.7, 15.8) (range, 2.9–23.3) months. Comparison of data collected from two institutions showed that there was no significant difference in age, gender, smoking history, tumor type, TNM stage, outcomes, etc. (P>0.05 for all).
Table 1
| Characteristics | Total number of patients (n=179) | Database 1 (n=133) | Database 2 (n=46) | P |
|---|---|---|---|---|
| Gender, n (%) | 0.13 | |||
| Male | 149 (83.2) | 114 (85.7) | 35 (76.1) | |
| Female | 30 (16.8) | 19 (14.3) | 11 (23.9) | |
| Age (years), median [IQR] | 63 [56, 67] | 63 [56, 67] | 62.5 [56.8, 68] | 0.93 |
| History of smoking, n (%) | 96 (53.6) | 74 (55.6) | 22 (47.8) | 0.36 |
| Histology, n (%) | 0.17 | |||
| Adenocarcinoma | 108 (60.3) | 75 (56.4) | 33 (71.7) | |
| Squamous cell carcinoma | 63 (35.2) | 51 (38.3) | 12 (26.1) | |
| Others | 8 (4.5) | 7 (5.3) | 1 (2.2) | |
| TNM stage, n (%) | 0.67 | |||
| III | 63 (35.2) | 48 (36.1) | 15 (32.6) | |
| IV | 116 (64.8) | 85 (63.9) | 31 (67.4) | |
| PD-1 expression, n/N (%) | 0.24 | |||
| Positive | 51/68 (75.0) | 28/40 (70.0) | 23/28 (82.1) | |
| Low | 18/68 (26.5) | 12/40 (30.0) | 6/28 (21.4) | |
| High | 33/68 (48.5) | 16/40 (40.0) | 17/28 (60.7) | |
| Negative | 17/68 (25.0) | 12/40 (30.0) | 5/28 (17.9) | |
| KRAS, n/N (%) | 0.82 | |||
| Positive | 11/71 (15.5) | 6/41 (14.6) | 5/30 (16.7) | |
| Negative | 60/71 (84.5) | 35/41 (85.4) | 25/30 (83.3) | |
| EGFR, n/N (%) | 0.56 | |||
| Negative | 104/115 (90.4) | 71/80 (88.8) | 33/35 (94.3) | |
| Positive | 11/115 (9.6) | 9/80 (11.3) | 2/35 (5.7) | |
| ALK, n/N (%) | >0.99 | |||
| Negative | 77/80 (96.3) | 66/69 (95.7) | 11/11 (100.0) | |
| Positive | 3/80 (3.8) | 3/69 (4.3) | 0/11 (0.0) | |
| ICI regimen, n (%) | ||||
| Anti PD-1 | 161 (89.9) | 118 (88.7) | 43 (93.5) | |
| nivolumab | 21 (11.7) | 14 (10.5) | 7 (15.2) | |
| Pembrolizumab | 23 (12.8) | 10 (7.5) | 13 (28.3) | |
| Camrelizumab | 21 (11.7) | 13 (9.8) | 8 (17.4) | |
| Tislelizumab | 38 (21.2) | 35 (26.3) | 3 (6.5) | |
| Toripalimab | 39 (21.8) | 38 (23.6) | 1 (2.2) | |
| Sintilimab | 14 (7.8) | 7 (5.3) | 7 (15.2) | |
| Bevacizumab | 5 (2.8) | 1 (0.8) | 4 (8.7) | |
| Anti PD-L1 | 18 (10.1) | 15 (11.3) | 3 (6.5) | |
| Atezolizumab | 8 (4.5) | 6 (4.5) | 2 (4.3) | |
| Durvalumab | 10 (5.6) | 9 (6.8) | 1 (2.2) | |
| Therapeutic strategy, n (%) | 0.16 | |||
| Monotherapy | 66 (36.9) | 45 (33.8) | 21 (45.7) | |
| Combination chemotherapy | 113 (63.1) | 88 (66.2) | 25 (54.3) | |
| PD at follow-up CT, n (%) | 0.70 | |||
| Yes | 50 (27.9) | 36 (27.1) | 14 (30.4) | |
| No | 129 (72.1) | 97 (72.9) | 32 (69.6) | |
| Median PFS (months), median [IQR] | 7.0 [3.4, 9.1] | 7.1 [3.4, 9.2] | 5.5 [3.4, 9.1] | 0.70 |
| Death, n (%) | 0.64 | |||
| Yes | 91 (50.8) | 69 (51.9) | 22 (47.8) | |
| No | 88 (49.2) | 64 (48.1) | 24 (52.2) | |
| Median follow-up time (months), median [IQR] | 23.9 [11.9, 24] | 23.3 [11.6, 24] | 24 [12.6, 24] | 0.51 |
| Median OS (months), median [IQR] | 12 [8.7, 15.8] | 11.7 [8.4, 15.9] | 12.4 [9.1, 17.2] | 0.54 |
IQR, interquartile range; TNM, tumor-node-metastasis; PD-1, programmed cell death protein 1; KRAS, Kirsten rat sarcoma viral oncogene homolog; EGFR, epidermal growth factor receptor; ALK, anapastic lymphoma kinase; ICI, immune checkpoint inhibitor; PD-L1, programmed death ligand-1; PD, progressive disease; CT, computed tomography; PFS, progression-free survival; OS, overall survival.
Clinical models established based on patients’ clinical characteristics to predict PFS and OS
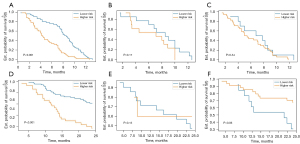
Age, gender, smoking history, tumor type, drugs used in ICI therapy, and TNM stage were selected for predicting outcomes in the clinical model (Table 2). Based on the clinical features, the model used for predicting PFS had the C-index of 0.631 in the training dataset; besides, the C-index was 0.540 and 0.543 in the validation and testing datasets, respectively. Meanwhile, for OS, the clinical model yielded C-index values of 0.674, 0.512, and 0.507 in the training, validation, and testing datasets, respectively. Furthermore, clinical features did not substantially affect the patients’ stratification by the Kaplan-Meier curves (P>0.05 for all, Figure S1). The results of time-independent ROC curve analysis of the three datasets are displayed in Figure S2.
Table 2
| Variables | Estimated | Standard error | t | Pr(>|z|) |
|---|---|---|---|---|
| PFS | ||||
| Gender | −0.8699 | 0.3272 | −2.6587 | 0.0078 |
| Age (years old) | −0.0115 | 0.0162 | −0.7119 | 0.4765 |
| Tumor type | 0.0207 | 0.1917 | 0.1079 | 0.9141 |
| Smoking status | 0.1526 | 0.2472 | 0.6176 | 0.5368 |
| TNM | 0.6340 | 0.2417 | 2.6231 | 0.0087 |
| Drug usage | 0.7010 | 0.3306 | 2.1207 | 0.0339 |
| OS | ||||
| Gender | −0.9705 | 0.3874 | −2.5051 | 0.0122 |
| Age (years old) | −0.0144 | 0.0194 | −0.7467 | 0.4553 |
| Tumor type | 0.0552 | 0.2362 | 0.2338 | 0.8151 |
| Smoking status | 0.2558 | 0.3210 | 0.7967 | 0.4256 |
| TNM | 0.69 | 0.3199 | 2.1564 | 0.0311 |
| Drug usage | −0.6388 | 0.5340 | −1.1963 | 0.2316 |
PFS, progression-free survival; TNM, tumor-node-metastasis; OS, overall survival.
Pre-treatment radiomics model for predicting PFS and OS
There were 76 features with poor repeatability (ICC <0.8) and 816 redundant features (r>0.8) removed. The remaining 145 radiomics features were selected by LASSO for further assessment. Then, six most contributing radiomic features were selected to establish a pre-treatment radiomics models by Cox PHs regression for predicting OS. Features contained in the radiomics model and their coefficients are summarized in Table 3. The radiomics model resulted in a moderate C-index value in the training dataset, while low values of the C-index were noted in both validation (0.558) and testing (0.558) datasets. Regarding PFS, eight radiomics features were selected based on the results of LASSO penalized logistic regression analysis (wavelet_HHL_firstorder_Mean, gradient_firstorder_Minimum, wavelet_LLL_glcm_ClusterProminence, gradient_glszm_GrayLevelVariance, wavelet_LLL_gldm_SmallDependenceHighGrayLevelEmphasis log_sigma_6_0_mm_3D_glcm_ClusterProminence, wavelet_LHL_glcm_ClusterShade log_sigma_6_0_mm_3D_gldm_HighGrayLevelEmphasis). The proposed model yielded C-index values of 0.704, 0.514, and 0.574 in the training, validation, and testing datasets, respectively. As shown in Figure 4, patients’ stratification was significant in the training dataset (P<0.001), while it was not significant in the validation and testing datasets (P>0.05). The results of time-independent ROC curve analysis of the three datasets are shown in Figure S3.
Table 3
| Variables | Estimated | Standard error | t | Pr(>|z|) |
|---|---|---|---|---|
| PFS | ||||
| (Intercept) | ||||
| Wavelet_HHL_firstorder_Mean | 0.1567 | 0.1079 | 1.4526 | 0.1463 |
| Gradient_firstorder_Minimum | 0.1337 | 0.1130 | 1.1834 | 0.2366 |
| Wavelet_LLL_glcm_ClusterProminence | 0.2188 | 0.1174 | 1.8643 | 0.0623 |
| Gradient_glszm_GrayLevelVariance | −0.1220 | 0.1223 | −0.9973 | 0.3186 |
| Wavelet_LLL_gldm_SmallDependenceHighGrayLevelEmphasis | 0.1704 | 0.1376 | 1.2377 | 0.2158 |
| Log_sigma_6_0_mm_3D_glcm_ClusterProminence | −0.0382 | 0.1916 | −0.1994 | 0.8420 |
| Wavelet_LHL_glcm_ClusterShade | 0.2053 | 0.1295 | 1.5849 | 0.1130 |
| Log_sigma_6_0_mm_3D_gldm_HighGrayLevelEmphasis | 0.4615 | 0.1986 | 2.3233 | 0.0202 |
| OS | ||||
| (Intercept) | ||||
| Wavelet_HHL_ngtdm_Contrast | 0.2004 | 0.1159 | 1.7292 | 0.0838 |
| Wavelet_HHL_firstorder_Maximum | −0.0371 | 0.1174 | −0.3160 | 0.7520 |
| Log_sigma_6_0_mm_3D_gldm_HighGrayLevelEmphasis | 0.3738 | 0.1108 | 3.3748 | 0.0007 |
| Wavelet_HHH_glcm_MCC | −0.0576 | 0.1071 | −0.5379 | 0.5906 |
| Log_sigma_6_0_mm_3D_glszm_SizeZoneNonUniformityNormalized | −0.1840 | 0.1115 | −1.6502 | 0.0989 |
| Wavelet_HLH_firstorder_Mean | −0.0196 | 0.1157 | −0.1694 | 0.8655 |
PFS, progression-free survival; OS, overall survival.
Delta-radiomics features for prediction of PFS and OS
According to the results of Pearson’s correlation analysis, 188 delta-radiomics features were selected to establish the predictive model. For PFS, five most contributing radiomic features delta-radiomics features (delta_wavelet_LHL_firstorder_Median, delta_original_shape_LeastAxisLength, delta_wavelet_LLL_glcm_Contrast, delta_wavelet_HLL_glcm_Idmn, delta_original_glcm_DifferenceAverage) were imported into the final model by Cox proportional hazards regression.
The model established based on delta-radiomics features yielded C-index values of 0.708, 0.688, and 0.603 in the training, validation, and testing datasets, respectively. For OS, seven most contributing delta-radiomics features (delta_wavelet_LHL_glcm_Imc2, delta_wavelet_HLH_firstorder_Median, delta_gradient_gldm_SmallDependenceLowGrayLevelEmphasis, delta_gradient_glszm_LowGrayLevelZoneEmphasis, delta_original_shape_LeastAxisLength, delta_original_glcm_DifferenceAverage) were selected based on the results of LASSO penalized logistic regression analysis. The selected features and corresponding coefficients are listed in Table 4. Then the seven most contributing delta-radiomics features were used to establish the delta radiomics models by Cox proportional hazards regression for predicting OS. A good prognostic performance was validated with the corresponding C-index values of 0.810, 0.762, and 0.697 in the training, validation, and testing datasets, respectively (Table 5). Furthermore, time-independent ROC curve analysis also confirmed that the delta-radiomics model had a good prognostic capability in the prediction of OS (Figure 5), and Kaplan-Meier curves showed that the delta-radiomics features could substantially affect the patients’ stratification (Figure 6).
Table 4
| Variables | Estimated | Standard error | t | Pr(>|z|) |
|---|---|---|---|---|
| PFS | ||||
| (Intercept) | ||||
| Delta_wavelet_LHL_firstorder_Median | 0.1746 | 0.1328 | 1.3151 | 0.1885 |
| Delta_original_shape_LeastAxisLength | 0.3624 | 0.1840 | 1.9697 | 0.0489 |
| Delta_wavelet_LLL_glcm_Contrast | −0.1647 | 0.1839 | −0.8957 | 0.3704 |
| Delta_wavelet_HLL_glcm_Idmn | 0.2326 | 0.1271 | 1.8298 | 0.0673 |
| Delta_original_glcm_DifferenceAverage | 0.1131 | 0.1541 | 0.7342 | 0.4629 |
| OS | ||||
| (Intercept) | ||||
| Delta_wavelet_LHL_glcm_Imc2 | −0.1162 | 0.1605 | −0.7242 | 0.4689 |
| Delta_wavelet_HLH_firstorder_Median | 0.3478 | 0.1347 | 2.5821 | 0.0098 |
| Delta_gradient_gldm_SmallDependenceLowGrayLevelEmphasis | 0.0810 | 0.1662 | 0.4872 | 0.6261 |
| Delta_gradient_glszm_LowGrayLevelZoneEmphasis | 0.1737 | 0.1930 | 0.8997 | 0.3683 |
| Delta_original_shape_LeastAxisLength | 0.1614 | 0.1443 | 1.1181 | 0.2635 |
| Delta_original_glcm_DifferenceAverage | −0.2092 | 0.1735 | −1.2059 | 0.2279 |
PFS, progression-free survival; OS, overall survival.
Table 5
| Models | Databases | PFS | OS | |||||
|---|---|---|---|---|---|---|---|---|
| C-index | AUC | Log-rank test P value | C-index | AUC | Log-rank test P value | |||
| Model 1 | Train | 0.631 | 0.668 | 0.25 | 0.675 | 0.735 | 0.65 | |
| Validation | 0.540 | 0.471 | 0.74 | 0.512 | 0.526 | 0.44 | ||
| Test | 0.543 | 0.501 | 0.84 | 0.507 | 0.580 | 0.55 | ||
| Model 2 | Train | 0.704 | 0.785 | <0.001 | 0.717 | 0.756 | <0.001 | |
| Validation | 0.514 | 0.541 | 0.17 | 0.571 | 0.632 | 0.15 | ||
| Test | 0.574 | 0.591 | 0.34 | 0.503 | 0.639 | 0.06 | ||
| Model 3 | Train | 0.708 | 0.787 | <0.001 | 0.810 | 0.898 | <0.001 | |
| Validation | 0.688 | 0.777 | 0.042 | 0.762 | 0.792 | 0.04 | ||
| Test | 0.603 | 0.608 | 0.03 | 0.697 | 0.716 | 0.02 | ||
| Model 4 | Train | 0.717 | 0.810 | 0.12 | 0.844 | 0.928 | <0.05 | |
| Validation | 0.683 | 0.751 | 0.42 | 0.709 | 0.811 | 0.15 | ||
| Test | 0.617 | 0.591 | 0.40 | 0.612 | 0.731 | 0.61 | ||
Model 1, clinical model; Model 2, pre-treatment radiomics model; Model 3, delta radiomics model; Model 4, mix model based on selected clinical and delta radiomics features. PFS, progression-free survival; OS, overall survival; C-index, concordance index; AUC, area under the curve.
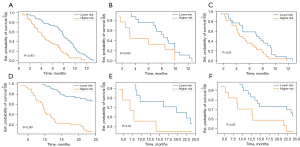
The mix model was developed with the selected clinical-CT and delta-radiomic features, and showed C-index values of 0.717, 0.683, and 0.617 for predicting PFS in the training, validation, and testing cohorts, respectively. To predict OS, the mix model yielded C-index values of 0.844, 0.709, and 0.612 in the three datasets. The predictive performance of mix model and delta-radiomics model were comparable (Table 5). Features contained in mix model and their coefficients are listed in Table 6.
Table 6
| Variables | Estimated | Standard error | t | Pr(>|z|) |
|---|---|---|---|---|
| PFS | ||||
| (Intercept) | ||||
| Delta_wavelet_LHL_firstorder_Median | 0.5369 | 0.2253 | 2.3832 | 0.0172 |
| Delta_original_shape_LeastAxisLength | 0.4903 | 0.2744 | 1.7869 | 0.9536 |
| Delta_wavelet_LLL_glcm_Contrast | −0.0587 | 0.3590 | −0.1634 | 0.8702 |
| Delta_wavelet_HLL_glcm_Idmn | 0.0496 | 0.1941 | 0.2555 | 0.7983 |
| Delta_original_glcm_DifferenceAverage | −0.3339 | 0.3072 | −1.0870 | 0.2771 |
| Gender | −0.2858 | 0.1542 | −1.8532 | 0.0639 |
| Age | −0.0481 | 0.1495 | −0.3220 | 0.7475 |
| Smoking | −0.0195 | 0.1786 | −0.1092 | 0.9131 |
| Durg usage | −0.1653 | 0.1845 | 0.8958 | 0.3704 |
| Tumor_type | 0.0061 | 0.1497 | 0.0406 | 0.9676 |
| TNM | 0.4453 | 0.2067 | 2.1545 | 0.0312 |
| OS | ||||
| (Intercept) | ||||
| Delta_wavelet_LHL_glcm_Imc2 | −0.3587 | 0.2743 | 0.6681 | 0.5041 |
| Delta_wavelet_HLH_firstorder_Median | 0.2938 | 0.2757 | 1.0656 | 0.2866 |
| Delta_gradient_gldm_SmallDependenceLowGrayLevelEmphasis | 0.5836 | 0.3110 | 1.8764 | 0.0606 |
| Delta_gradient_glszm_LowGrayLevelZoneEmphasis | 0.1737 | 0.1930 | 0.8997 | 0.3683 |
| Delta_original_shape_LeastAxisLength | 0.3187 | 0.2588 | 1.2311 | 0.2183 |
| Delta_original_glcm_DifferenceAverage | −0.2993 | 0.2523 | −1.1863 | 0.2355 |
| Gender | −0.5613 | 0.1710 | −3.2830 | 0.0010 |
| Age | −0.2739 | 0.1562 | −1.7537 | 0.8954 |
| Smoking | 0.0239 | 0.1816 | 0.1315 | 0.1315 |
| Durg usage | −0.2036 | 0.1787 | −1.1391 | 0.2546 |
| Tumor_type | 0.2365 | 0.1548 | 1.5272 | 0.1267 |
| TNM | −0.0395 | 0.1756 | −0.2252 | 0.8218 |
PFS, progression-free survival; TNM, tumor-node-metastasis; OS, overall survival.
Discussion
The present study introduced a delta-radiomics model demonstrating strong performance in predicting response to ICI therapy and patients’ prognosis. Notably, this model surpassed both the clinical model and the pre-treatment radiomics model, which were established based on patients’ demographic information and pre-treatment CT radiomics features, respectively. Moreover, these findings were further confirmed by an external independent database.
Based on the findings of the present study, two delta-radiomics features were chosen to predict both PFS and OS in the delta-radiomics model. Specifically, (I) the delta shape_LeastAxisLength denoted alterations in the smallest axis length of the ROI-enclosing ellipsoid. This selection appeared rational, considering that changes in tumor diameter typically serve as sensitive indicators of treatment response, aligning with the iRECIST guidelines concerning alterations in tumor size. (II) Firstorder_Median reflected changes of the median gray-level intensity within the tumor. A recent study (27) demonstrated the firstorder_Median parameter observed on enhanced T1-weighted magnetic resonance (MR) sequences has been demonstrated to correlate with immunophenotyping and serves as a radiomics biomarker for predicting OS in patients diagnosed with intrahepatic cholangiocarcinoma. We assumed that the mentioned outcome-related feature might be correlated to the malignant biological behavior of the tumor. Besides, all the changes in other radiomics features included in the delta-radiomics model were gray-level features, such as GLCM, GLDM, and GLSZM, which reflected the changes in tumor heterogeneity due to ICI treatments. Prior studies have indicated that tumor heterogeneity serves as a predictor of OS in NSCLC patients (28,29) or immune response (30). Although the precise mechanism remains unclear, tumor heterogeneity may signify clonal dominance or genomic diversity within tumors, which has been identified as a primary factor contributing to therapeutic resistance (31). Another consideration is that variations in tumor heterogeneity could be linked to differing densities and distributions of CD8+ T cells, the predominant effector cell type in cancer and cancer immunotherapy (27,32). Furthermore, the radiomic signature of CD8+ T cells has been proven to predict the immunophenotype of solid tumors and clinical outcomes in patients with cancer who had been treated with ICI therapy (33). Therefore, it is reasonable to hypothesize that these three radiomics biomarkers, which reflect changes in tumor size, density, and heterogeneity, hold predictive value for prognosis and overall response to immunotherapy within the quantitative CT radiomics group. Consolidating these biomarkers into a signature could potentially offer more substantial insights for guiding dependable clinical decisions. Khorrami et al. (8) applied changes in the radiomic texture (DelRADx) to predict response to immunotherapy and explored the association of delta-radiomic risk-score (DRS) with OS in NSCLC. They also found that the DelRADx could be used to distinguish responders from non-responders with a good performance [area under the curve (AUC), 0.88±0.08]. Besides, DRS was found to be a biomarker for predicting OS (C-index =0.72), which was in agreement with our results and validated the predictive value of the delta-radiomics model in immunotherapy. In addition to establishment of a delta-radiomics model and assessment of its capabilities, we simultaneously developed the clinical model and baseline radiomics model to compare their predictive values. This is important because delta radiomics could be introduced into the routine clinical workflow if it provides higher predictive value than clinical factors or the radiomics features extracted from pre-treatment CT data. In addition, Khorrami et al. (8) extracted delta-radiomics features from contrast-enhanced CT images, while those features in our study were obtained from non-contrast-enhanced CT images. However, contrast agents may obscure imaging features that reflect potential tumor heterogeneity (34), and the heterogeneity of contrast-enhanced protocols, e.g., different dosage and duration of dosing, could affect the reproducibility of the radiomics.
As anticipated, the proposed delta-radiomics model demonstrated superior performance in predicting the prognosis of NSCLC patients. This superiority can be attributed to several factors, as elaborated below: First, among the clinical features, TNM staging showed the highest correlation coefficient (0.69) among the selected features for the prediction of OS. However, as it was developed based on tumor size, lymph node metastasis, and metastasis status, it only provided anatomical data. Patients with even the same tumor stage might have different levels of prognosis (35). In contrast, radiomics captures tumor characteristics from medical images, offering a robust method for interpreting intratumor heterogeneity, a capability lacking in traditional tumor staging. Additionally, the delta-radiomics model can evaluate the relative net change of radiomics features across longitudinal images. This capacity not only enables the identification and quantification of therapy-induced changes but also holds potential for predicting such changes over the treatment duration (36). Moreover, the study suggests that treatment-induced changes in the radiomic profile have the potential to enhance the predictive accuracy of OS or disease-free survival (DFS) in cancer patients (37,38). However, Fave et al. (17) explored the utility of delta-radiomics features in predicting clinical outcomes for NSCLC patients undergoing radiation therapy. Their findings revealed that incorporating delta-radiomics features into pre-treatment models significantly improved the C-index value (from 0.672 to 0.675). However, this addition did not notably impact patient stratification, as demonstrated by Kaplan-Meier curves. Therefore, further research is necessary to validate the application of delta-radiomics features in predicting patients’ prognosis.
The limitations of the present study should be pointed out. First, a small sample size restricted the generalization of our findings. Second, due to the retrospective nature of the study, some important clinical data were missing [e.g., the expression levels of PD-1 and epidermal growth factor receptor (EGFR)]. Third, owing to the relatively short-term follow-up, long-term follow-up should be conducted to more reliably predict OS. Fourth, the independent test set consistently yields lower results compared to the validation data in our models, likely due to the use of CT scanning machines from different manufacturers at the two centers. Moreover, given the intricate nature of the survival analysis model and our limited sample size, variations in performance were observed across the training, test, and validation sets. Nevertheless, the C-index consistently fell within the range established by fivefold cross-validation on the training set, indicating overall stability of our model. Last but not least, as the images used in our study were non-contrast-enhanced CT images, the vessels that passed across lesions could not be segmented. Thus, the tumor ROI might contain blood vessels of solid tumor components of interest, which might influence radiomics characteristics and the calculated tumor volume.
Conclusions
In summary, the proposed delta-radiomics model demonstrated promising performance in predicting clinical outcomes among advanced NSCLC patients undergoing ICI therapy. Moreover, it surpassed both the clinical and pre-treatment radiomics models, underscoring the importance of delta-radiomics features in forecasting immunotherapeutic response in this patient population. Nevertheless, further research is warranted to address the aforementioned limitations and validate our findings.
Acknowledgments
We would like to thank all colleagues for helping us during the current study.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-7/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-7/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-7/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-7/coif). C.D. is an employee of Bayer Healthcare. X.Z. is an employee of Philips Healthcare. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval for this retrospective study was obtained from the Ethics Committee of Wuhan Union Hospital (Wuhan, China) (Approval No. S0516), with the requirement for written informed consent waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Okazaki T, Chikuma S, Iwai Y, et al. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol 2013;14:1212-8. [Crossref] [PubMed]
- Sharma P, Goswami S, Raychaudhuri D, et al. Immune checkpoint therapy-current perspectives and future directions. Cell 2023;186:1652-69. [Crossref] [PubMed]
- Mountzios G, Remon J, Hendriks LEL, et al. Immune-checkpoint inhibition for resectable non-small-cell lung cancer - opportunities and challenges. Nat Rev Clin Oncol 2023;20:664-77. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Mulkey F, Theoret MR, Keegan P, et al. Comparison of iRECIST versus RECIST V.1.1 in patients treated with an anti-PD-1 or PD-L1 antibody: pooled FDA analysis. J Immunother Cancer 2020;8:e000146. [Crossref] [PubMed]
- Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol 2017;18:e143-52. [Crossref] [PubMed]
- Win T, Miles KA, Janes SM, et al. Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer. Clin Cancer Res 2013;19:3591-9. [Crossref] [PubMed]
- Khorrami M, Prasanna P, Gupta A, et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non-Small Cell Lung Cancer. Cancer Immunol Res 2020;8:108-19. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Zhang G, Cao Y, Zhang J, et al. Predicting EGFR mutation status in lung adenocarcinoma: development and validation of a computed tomography-based radiomics signature. Am J Cancer Res 2021;11:546-60. [PubMed]
- Song L, Zhu Z, Mao L, et al. Clinical, Conventional CT and Radiomic Feature-Based Machine Learning Models for Predicting ALK Rearrangement Status in Lung Adenocarcinoma Patients. Front Oncol 2020;10:369. [Crossref] [PubMed]
- Gu Y, She Y, Xie D, et al. A Texture Analysis-Based Prediction Model for Lymph Node Metastasis in Stage IA Lung Adenocarcinoma. Ann Thorac Surg 2018;106:214-20. [Crossref] [PubMed]
- Coroller TP, Agrawal V, Huynh E, et al. Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC. J Thorac Oncol 2017;12:467-76. [Crossref] [PubMed]
- Chen M, Copley SJ, Viola P, et al. Radiomics and artificial intelligence for precision medicine in lung cancer treatment. Semin Cancer Biol 2023;93:97-113. [Crossref] [PubMed]
- Lee G, Park H, Sohn I, et al. Comprehensive Computed Tomography Radiomics Analysis of Lung Adenocarcinoma for Prognostication. Oncologist 2018;23:806-13. [Crossref] [PubMed]
- Ma Y, Ma W, Xu X, et al. How Does the Delta-Radiomics Better Differentiate Pre-Invasive GGNs From Invasive GGNs? Front Oncol 2020;10:1017. [Crossref] [PubMed]
- Fave X, Zhang L, Yang J, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep 2017;7:588. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Ligero M, Garcia-Ruiz A, Viaplana C, et al. A CT-based Radiomics Signature Is Associated with Response to Immune Checkpoint Inhibitors in Advanced Solid Tumors. Radiology 2021;299:109-19. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Pölsterl S. scikit-survival: A Library for Time-to-Event Analysis Built on Top of scikit-learn. J Mach Learn Res 2020;21:1-6.
- van der Walt S, Schönberger JL, Nunez-Iglesias J, et al. scikit-image: image processing in Python. PeerJ 2014;2:e453. [Crossref] [PubMed]
- Lv L, Xin B, Hao Y, et al. Radiomic analysis for predicting prognosis of colorectal cancer from preoperative (18)F-FDG PET/CT. J Transl Med 2022;20:66. [Crossref] [PubMed]
- Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med 2016;15:155-63. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Ortega C, Eshet Y, Prica A, et al. Combination of FDG PET/CT Radiomics and Clinical Parameters for Outcome Prediction in Patients with Hodgkin's Lymphoma. Cancers (Basel) 2023;15:2056. [Crossref] [PubMed]
- Zhang J, Wu Z, Zhao J, et al. Intrahepatic cholangiocarcinoma: MRI texture signature as predictive biomarkers of immunophenotyping and survival. Eur Radiol 2021;31:3661-72. [Crossref] [PubMed]
- Trebeschi S, Kurilova I, Călin AM, et al. Radiomic biomarkers for the prediction of immunotherapy outcome in patients with metastatic non-small cell lung cancer. J Clin Oncol 2017;25:e14520. [Crossref]
- Grove O, Berglund AE, Schabath MB, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One 2015;10:e0118261. [Crossref] [PubMed]
- Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017;6:e23421. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Raskov H, Orhan A, Christensen JP, et al. Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br J Cancer 2021;124:359-67. [Crossref] [PubMed]
- Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. [Crossref] [PubMed]
- Yip SSF, Liu Y, Parmar C, et al. Associations between radiologist-defined semantic and automatically computed radiomic features in non-small cell lung cancer. Sci Rep 2017;7:3519. [Crossref] [PubMed]
- Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 2020;20:662-80. [Crossref] [PubMed]
- Nasief H, Zheng C, Schott D, et al. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. NPJ Precis Oncol 2019;3:25. [Crossref] [PubMed]
- Fave X, Zhang L, Yang J, et al. TU-D-207B-02: Delta-Radiomics: the prognostic value of therapy-induced changes in Radiomics features for stage III non-small cell lung cancer patients. Med Phys 2016;43:3750. [Crossref]
- Jeon SH, Song C, Chie EK, et al. Delta-radiomics signature predicts treatment outcomes after preoperative chemoradiotherapy and surgery in rectal cancer. Radiat Oncol 2019;14:43. [Crossref] [PubMed]


