
PSME3 promotes lung adenocarcinoma development by regulating the TGF-β/SMAD signaling pathway
Highlight box
Key findings
• Proteasome activator subunit 3 (PSME3) expression was abnormally elevated in lung adenocarcinoma (LUAD), and patients with a high expression of PSME3 had a poor prognosis. PSME3 promoted LUAD cell proliferation, migration, and invasion, accelerated cell cycle progression, and inhibited apoptosis. PSME3 also regulated the transforming growth factor-beta (TGF-β)/SMAD signaling pathway to promote LUAD progression.
What is known and what is new?
• PSME3 is involved in the development of diseases, such as neurological disorders, cardiovascular diseases, immune-related diseases, and many types of cancers. However, prior to this study, its role in LUAD was still unknown.
• In this study, we demonstrated for the first time that the expression of PSME3 is elevated in LUAD, and PSME3 promotes LUAD progression through the TGF-β/SMAD signaling pathway.
What is the implication, and what should change now?
• The experimental results demonstrated that PSME3 is a key factor and could serve as a novel target in LUAD. Our results support the targeting of PSME3 as a new therapeutic approach for the clinical treatment of patients with LUAD.
Introduction
In the last few years, cancer-related deaths have been predominantly attributed to lung cancer, which is one of the most prevalent malignant tumors in the world (1,2). As the most common histological subtype of lung cancer, patients with lung adenocarcinoma (LUAD) tend to have a poor prognosis (3). With the development of medical technology, the advent of molecularly targeted therapies and immunotherapies has yielded enormous benefits for patients with LUAD. Therefore, the identification of a new molecular target is important for the treatment of patients with LUAD and is urgently needed.
The proteasome is a ubiquitin-dependent giant protein complex with multiple protein hydrolase activities, and consists of multiple subunits with different functions (4,5). The ubiquitin-proteasome system (UPS) regulates a variety of biological processes, including gene expression, oxidative stress, immune escape, cell apoptosis, and the cell cycle (6). The UPS has been found to affect a variety of diseases, including neurodegenerative (7,8), cardiovascular (9), immunity-related diseases (10,11), and cancer (12). Proteasome inhibitors have been used in a wide variety of applications (13), including the treatment of systemic lupus erythematosus (14), multiple myeloma (15), and breast cancer (16). Therefore, studies of the proteasome and its inhibitors in LUAD could yield very promising results in relation to the treatment of patients with this cancer.
Proteasome activator subunit 3 (PSME3) is located on chromosome 17q21.31. The exact function and mechanism of PSME3 have not yet been fully clarified; however, studies have revealed that PSME3 regulates the degradation of the cell cycle inhibitor p21 (17,18), promotes the degradation of SRC-3 proteins by ubiquitin- and adenosine 5'-triphosphate (ATP)-independent means (19), and drives the degradation of the tumor suppressor p53 (20). There is an association between PSME3 expression and the presentation of different major histocompatibility complex class I (MHC-I) antigenic peptides, and PSME3 plays a crucial role in processing different protein tyrosine phosphatases (PTPs) and negatively regulating cluster of differentiation CD8+ T cell responses to cancer (21). PSME3 also participates in the inhibition of the aggregation of disease-related proteins at low temperatures (22). In recent years, a PSME3 interactor, PIP30, has been identified that binds with high specificity to PSME3 and enhances its binding to the cellular 20S proteasome, affecting the specificity of PSME3 for peptide substrates (23). Recently, an increasing number of studies have found that PSME3 is strongly associated with cancers, including pancreatic cancer (24,25), skin melanoma (26), thyroid cancer (27,28), oral squamous cell carcinoma (29), esophageal cancer (30,31), gastric cancer (32), colorectal cancer (33-35), cervical cancer (36), and breast cancer (37). However, the function and mechanism of PSME3 in LUAD have not been found.
This study was designed to examine the expression, biological function, and potential mechanism of PSME3 in LUAD, and the results suggest that PSME3 is a feasible target for the treatment of patients with LUAD. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-340/rc).
Methods
Collection of clinical samples
Ten pairs of fresh LUAD and adjacent normal tissues were obtained from patients who underwent surgical resection at the Thoracic Surgery Department of Qilu Hospital. The samples were stored in liquid nitrogen until use. All diagnoses were confirmed by pathology. A protocol was prepared before the study without registration. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of Qilu Hospital (No. KYLL-2021[KS]-1053), and informed consent was obtained from each patient.
Cell culture, transfection, and selection
A549, H1299, PC9, and H1975 cell lines were provided by Shanghai Cell Bank Research Center, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle medium and RPMI-1640 medium (Gibco, NY, USA), containing 10% fetal bovine serum (FBS; BI, Beit Haemek, Israel), at 37 °C, and 5% carbon dioxide. Short tandem repeat (STR) was used to identify all the cell lines, and the cultures were regularly tested for mycoplasma contamination every three months. The cells were transiently transfected with small-interfering RNA or plasmid using jetPRIME transfection reagent (Polyplus, NY, USA) in accordance with the manufacturer’s instructions. Approximately 48 hours after transfection, the cells were collected and lysed to assess transfection efficiency. PSME3 overexpression and knockdown lentiviruses were purchased from Jikai Company (Shanghai, China). The A549 cells were transfected with the PSME3 knockdown lentivirus [multiplicity of infection (MOI) =10], and the PC9 cells were transfected with the PSME3 overexpression lentivirus (MOI =20). We performed a polyclonal selection and the cells were cultured with 0.5 mg/mL of puromycin 48 hours after transfection for one week to obtain the stable expressing cells.
RNA isolation, cDNA preparation, and quantitative real time polymerase chain reaction (qRT-PCR)
Total cellular RNA was extracted using the Rapid Cellular RNA Extraction Kit (AC0205-B; SparkJade, Jinan, China), and then reverse transcribed to complementary DNA using the Reverse Transcription Kit (AG11706; Accurate Biology, Changsha, China). qRT-PCR was performed on a LightCycler® 480 Instrument II detection system (Roche, Basel, Switzerland) using the SYBR Green Premix Pro Taq HS qPCR kit (AG11701; Accurate Biology, Changsha, China). GenePharma company (Suzhou, China) provided the following primers for PSME3: (F: 5'-TCAAAATGTGGGTACAGCTCCT-3', R: 5'-CTCCTGAATGGACACCCCAA-3'), and β-Tubulin (F: 5'-CATGGACTCTGTTCGCTCAGG-3', R: 5'-CCTTGGCCCAGTTGTTACCT-3'). The expression of the target genes was calculated using the 2−ΔΔCT method. To ensure accuracy and reliability, the experiment was replicated three times.
Western blotting (WB) assays
The cells and LUAD samples were lysed on ice in radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China) containing 1% protease and phosphatase inhibitors, for 30 minutes and vortexed every 10 minutes with shaking. The samples were centrifuged at 12,000 ×g and 4 °C for 15 minutes. The supernatant was then collected, and the protein concentration was calculated using a bicinchoninic acid assay kit (Beyotime, Shanghai, China). The total amount of protein per well was around 30 micrograms. The proteins were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. After blocking with 5% skim milk powder for 1 hour at room temperature, the membranes were incubated with primary antibody at 4 °C overnight. After washing three times with Tris Buffered Saline with Tween-20 (TBST), the membranes were incubated with secondary antibody for 1 hour at room temperature. Protein bands were then visualized via enhanced chemiluminescence in accordance with the manufacturer’s instructions. The following primary antibodies were used for this experiment: anti-PSME3 (14907-1-AP; Proteintech, Chicago, USA), anti-B-cell lymphoma-2 (anti-Bcl-2) (ET1702-53; HUABIO, Hangzhou, China), anti-Bcl-2-associated X (anti-BAX) (ER0907; HUABIO), anti-cyclin dependent kinase 4 (anti-Cdk4) (ET1612-23; HUABIO), anti-cyclin dependent kinase 6 (anti-Cdk6) (ET1612-3; HUABIO), anti-cyclin A1 + cyclin A2 (ET1612-8; HUABIO), anti-cyclin D1 (ET1601-31; HUABIO), anti-cyclin E1 (ET1612-16; HUABIO), anti-N-cadherin (22018-1-AP; Proteintech), anti-E-cadherin (A20798; Abclonal, Wuhan, China), anti-vimentin (ET1610-39; HUABIO), anti-snail family transcriptional repressor 2 (anti-SNAI2) (EM1706-65; HUABIO), anti-SNAIL (ER1706-22; HUABIO), anti-poly ADP-ribose polymerase (PARP1) and anti-cleaved PARP1 (13371-1-AP; Proteintech), anti-caspase 9 and anti-cleaved caspase 9 (10380-1-AP; Proteintech), anti-caspase 3 and anti-cleaved caspase 3 (19677-1-AP; Proteintech), anti-Smad2/3 (AF6367; Affinity Biosciences, Cincinnati, OH, USA), anti-p-Smad2/3 (AF3367; Affinity Biosciences), anti-transforming growth factor-beta (anti-TGF-β) receptor II (ER1917-66; HUABIO), and anti-beta Tubulin (AB0039; Abways, Shanghai, China). The experiments were independently conducted three times.
Tissue microarray, immunohistochemistry (IHC), and hematoxylin and eosin (H&E) staining
The tissue microarray (HlugA180Su11), which contained 174 available tissues, including 85 LUAD tissues and 89 adjacent tissues, was purchased from Shanghai Outdo Biotech (Shanghai, China). IHC and H&E staining were performed, and the IHC results were scored individually by two pathologists. The histochemical score (H-score) indicated the percentage of positive cells (ranging from 0% to 100%) and was assessed as follows: H score = percentage of positive cells × staining intensities (where 0 = none; 1 = weak; 2 = moderate; and 3 = strong). Based on the scoring results, X-title software was used to determine the optimal cut-off values for high and low expression. Images were captured with a microscope (Olympus, Tokyo, Japan).
Cell Counting Kit-8 (CCK-8) assays
The transfected cells were cultured in 96-well plates for 24, 48, 72, and 96 hours. CCK-8 (#K1018; APExBIO, Houston, USA) medium was added to the cells, and the cells were incubated for 2 hours in the dark. The absorbance of each well was measured at 450 nm with a microplate reader. This experiment was performed independently three times.
5-ethynyl-2'-deoxyuridine (EdU) assays
The transfected cells were incubated overnight in 96-well plates. EdU staining was carried out with an EdU staining kit (Beyotime, Shanghai, China) in accordance with the manufacturer’s instructions. Images were ultimately captured by fluorescence microscope (Olympus, Tokyo, Japan).
Colony formation experiment
We performed our experiments with single cell suspensions, inoculated with 800 cells per well in six-well plates. The transfected cells were cultured until colony formation was visible to the naked eye. The cells were fixed with 4% paraformaldehyde for 20 minutes, stained with 0.5% crystal violet for 30 minutes and washed three times with phosphate buffered saline (PBS). Images were captured with a digital camera. The experiment was repeated three times independently.
Flow cytometry
The cells were cultured in six-well plates, and cell cycle assays were performed by flow cytometry after 48 hours. The Cell Cycle Kit (Multi Sciences, Hangzhou, China) was used to detect the cell cycle in accordance with the manufacturers’ instructions.
Wound-healing assays
The cells were cultured in six-well plates with complete medium until the cells were nearly confluent. Next, a 200-µL pipette tip was used to make scratches, which were straight and the same width in all wells. After being washed three times with PBS, the cells were incubated with a serum-free medium. The wound area was photographed with a light microscope at 0 and 24 hours, and measured using ImageJ software (Germany). The experiment was repeated three times.
Transwell assays
These experiments were performed using transwell chambers in 24-well plates (8 μm; Corning, NY, USA). For the migration experiments, the LUAD cells incubated with different treatments were seeded in the upper chamber containing serum-free medium, and medium containing 20% FBS was added to the lower chamber. After 24 hours of incubation at 37 °C, the cells were gently removed from the upper surface of the cell membrane with a cotton swab. The migrated cells were fixed with 4% paraformaldehyde at room temperature for 30 minutes and stained with 0.1% crystal violet dye for 30 minutes. The invasion assay was performed with Matrigel in the upper chamber, the remaining procedure was the same as that for the migration assay. These experiments were performed three times independently.
In vivo experiment
Four-week-old female BLAB/c nude mice were purchased from GemPharmatech (Nanjing, China) and randomly assigned to cages. To ensure a comfortable environment, each cage housed five mice. The conditions under which nude mice grow were the same. To construct a subcutaneous xenograft tumor model, we injected stably transduced A549 and PC9 cells into the right axilla of the mice (5×106 per mouse) and recorded the size of the tumors on a weekly basis. Tumor volume was calculated as follows: volume = length × width2/2. After nearly four weeks, the mice were euthanized (the death of a mouse during the experiment was not counted), and the tumors were collected carefully and photographed with a digital camera. And the tumors were subsequently subjected to H&E staining and IHC. The animal experiments were performed under a project license (No. DWLL-2023-056) granted by the Animal Research Ethics Committee of Qilu Hospital, in compliance with institutional guidelines for the care and use of animals.
Data processing and transcriptome sequencing
PSME3 expression data were obtained from The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/) and the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) databases. Transcriptome RNA sequencing (RNA-seq) was performed by Hangzhou Kaitai Biotechnology Co., Ltd. (Hangzhou, China). Changes in the expression levels of the messenger RNA (mRNA) transcripts were determined by comparing control cells with A549 cells in which PSME3 was knocked down.
Statistical analysis
GraphPad Prism (GraphPad 8, USA), SPSS (version 25.0, USA), and R (version 4. 3.1, New Zeeland) software were used to analyze the data. The data are presented as the mean ± standard deviation. The Student’s t-test (two-tailed) was used to analyze the differences between two groups. One-way analysis of variance (ANOVA) was used to compare differences among more than two groups. The Kaplan-Meier survival analysis, Pearson Chi-squared test and Fisher exact test were performed as indicated. A P value <0.05 was considered statistically significant.
Results
PSME3 expression was significantly increased in LUAD and was associated with poor patient prognosis
First, we used the high-throughput sequencing data to compare PSME3 expression in the LUAD and non-cancerous lung tissues. In TCGA and GEO databases, we found that PSME3 expression was significantly higher in the LUAD tissues than the normal tissues (Figure 1A). The correlation between the clinical characteristics and PSME3 expression in LUAD patients was then analyzed using the GEPIA2 website (http://gepia2.cancer-pku.cn/), and a positive correlation with tumor stage was found (Figure 1B). We assessed the relationship between PSME3 expression and the prognosis of LUAD patients using the GEPIA2 website and the Kaplan-Meier survival analysis, and found that elevated PSME3 expression indicated a poorer patient prognosis (Figure 1C). We then performed an IHC analysis of the tissue microarrays, and the IHC scores indicated that PSME3 expression was much higher in the LUAD tissues than the corresponding adjacent normal lung tissues (Figure 1D,1E). Further, the Kaplan-Meier survival analysis showed that patients with high PSME3 expression tended to have a worse prognosis (Figure 1F). Next, WB assays were performed to analyze proteins extracted from 10 pairs of fresh tissues and adjacent normal lung tissues from patients with LUAD, and PSME3 expression was found to be significantly higher in the LUAD tissues than the normal tissues (Figure 1G). The above experiments confirmed that PSME3 was highly expressed in LUAD and was associated with poor patient prognosis.

PSME3 promoted LUAD cell proliferation in vivo and in vitro
First, the WB and qRT-PCR assay results showed that PSME3 expression was highest in the A549 cells and was lowest in the PC9 cells among four cell lines tested. Thus, we knocked down PSME3 in the A549 cells and overexpressed PSME3 in the PC9 cells. We then verified the efficiency of the knockdown and overexpression in the A549 and PC9 cells, respectively, by qRT-PCR and WB. The expression of PSME3 was significantly reduced in the A549 cells after knockdown, and significantly increased in the PC9 cells after overexpression (Figure 2A,2B). Subsequently, CCK-8 assays, EdU assays, and colony formation assays were performed to assess the proliferation ability of the transfected cells. We found that the proliferation ability of the cells significantly decreased after the knockdown of PSME3, and the proliferation ability of the cells significantly increased after the overexpression of PSME3 (Figure 2C-2G). To further verify the effect of the proliferative ability of PSME3 in vivo, we used BALB/c-nude strain mice for xenograft subcutaneous tumor-forming experiments. The tumor growth rate, tumor volume, and tumor weight were significantly lower in the PSME3 knockdown mice than the control mice; the opposite results were observed for the mice in the overexpression group (Figure 3A-3C). The tumors were then stained (by H&E and IHC). The expression of Ki-67 was significantly lower in the knockdown group than the control group; the opposite result was observed in the overexpression group (Figure 3D). The above experiments demonstrated that PSME3 promoted the proliferation of LUAD cells both in vivo and in vitro.


PSME3 promoted the migration and invasion of LUAD
To further investigate the effect of PSME3 on LUAD cell migration and invasion, we performed Transwell experiments and found that the number of A549 cells that migrated decreased significantly after the knockdown of PSME3; similarly, the number of PC9 cells that migrated increased significantly after the overexpression of PSME3. When the chambers were lined with Matrigel, the same above-mentioned trends were observed, except that the overall numbers were lower (Figure 4A,4B). Next, we performed wound-healing experiments. The results showed that the wound-healing area was significantly smaller in the PSME3 knockdown cells than the control cells. The wound-healing area was significantly larger in the PSME3 overexpressing cells than the control cells (Figure 4C,4D). Finally, we used WB to analyze the expression of some critical epithelial-mesenchymal transition (EMT) regulatory proteins. The results showed that the decreased expression of PSME3 led to the downregulation of N-cadherin, Vimentin, SNAI2, and Snail, and the upregulation of E-cadherin; the opposite results were observed after the overexpression of PSME3 (Figure 4E). Overall, the results of the above experiments demonstrated that PSME3 had the ability to promote the migration and invasion of LUAD cells.

PSME3 promoted cell cycle progression while inhibiting cell apoptosis
To further explore the role of PSME3 in LUAD, we analyzed PSME3 by flow cytometry. First, we found that the knockdown of PSME3 resulted in G1/S phase arrest and that the overexpression of PSME3 markedly accelerated the G1/S phase transition (Figure 5A,5B). In addition, we also examined the expression of some key biomarkers related to the cell cycle and apoptosis after PSME3 knockdown or overexpression. We found that after the knockdown of PSME3 in the A549 cells, the expression of BAX, cleaved PARP1, cleaved caspase 9, cleaved caspase 3 significantly increased, and the expression of Bcl-2, Cdk4, Cdk6, cyclin A1 + cyclin A2, cyclin D1 and cyclin E1 significantly decreased; the opposite results were observed after the overexpression of PSME3 in the PC9 cells (Figure 5C,5D). From the above experiments, we determined that PSME3 promoted cell proliferation by promoting cell cycle progression and inhibiting apoptosis.

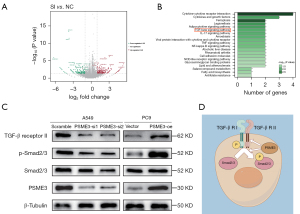
PSME3 promoted LUAD progression by activating the TGF-β/SMAD signaling pathway
Previous functional experiments demonstrated that PSME3 promoted LUAD; however, the exact mechanism involved has not yet been elucidated. To identify a clear pathway by which PSME3 affects the development of LUAD, we prepared A549 cells after the knockdown of PSME3 and control cells for transcriptome sequencing. We found that the expression of 431 genes was significantly altered, including 183 genes that were downregulated and 248 genes that were upregulated (Figure 6A). The expression levels of many molecules in the SMAD family were significantly altered, suggesting that the expression level of PSME3 affected the SMAD family. In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that PSME3 alterations affected some tumor and other signaling pathways, including the TGF-β signaling pathway (Figure 6B). The TGF-β pathway orchestrates many cellular processes, including cell growth, differentiation, migration and invasion, and extracellular matrix remodeling (38). The pathway and its components have been shown to be dysregulated in a wide variety of tumors; and members of the SMAD family are important downstream targets of this pathway; therefore, we chose to analyze the TGF-β/SMAD pathway. Then we analyzed the expression of TGF-β receptor II, Smad2/3, and p-Smad2/3. The WB results showed that the knockdown of PSME3 in the A549 cells decreased the expression of TGF-β receptor II and p-Smad2/3, and that the overexpression of PSME3 in the PC9 cells significantly increased the expression of TGF-β receptor II and p-Smad2/3 (Figure 6C). We then mapped the pattern of PSME3 affecting TGF-β signaling pathway by Figdraw, which demonstrated how PSME3 acted in the intracellular TGF-β signaling pathway (Figure 6D). Therefore, these results demonstrated that PSME3 activated the TGF-β/SMAD signaling pathway in the LUAD cells.
Discussion
In this study, we demonstrated for the first time that PSME3 positively regulates the development of LUAD cells, is associated with poor patient prognosis, and affects the TGF-β/SMAD signaling pathway. These findings suggest that PSME3 could serve as a new target in the clinical treatment of patients with LUAD.
PSME3 is an activator of the 20S proteasome that induces the hydrolysis of a variety of substrates in an ATP- and ubiquitin-independent manner (39). A recent study showed that PSME3 acts by binding to the 20S proteasome variant to activate the trypsin-like (T-L) protein hydrolysis site (40). Research has shown that PSME3 knockdown sensitizes multiple myeloma (MM) cells to bortezomib-mediated growth arrest, as sensitization inhibits proteasomal capacity (41). At the G1/S checkpoint, the p53 pathway regulates the induction of different phenotypes in a stochastic manner in a number of cell lines (42). In our experiments, we also found that PSME3 promoted the proliferation of LUAD cells and accelerated cell cycle progression. It has been reported that PSME3 knockdown inhibits melanoma cell growth and arrests melanoma cells in the G1 phase (43). In addition, PSME3 knockdown can enhance the radiosensitivity of colorectal cancer cells by triggering cell cycle arrest in the G1/M phase through the downregulation of cyclin B2 and CDK1 (34). Together, these findings suggest that PSME3 is involved in and plays an important role in regulating the cell cycle.
A previous study has shown that the proteasome activator PSME3 acts as an antiapoptotic regulator (44). In our experiments, we demonstrated for the first time that PSME3 overexpression inhibited LUAD cell apoptosis and that PSME3 knockdown had the opposite effect. Another study has shown that miR-7-5p inhibits cell proliferation and induces apoptosis in breast cancer cells mainly by targeting PSME3 (45). A new quinolone analog, LZ-106, was recently found to be involved in the generation of reactive oxygen species (ROS) in activated cells upon P53 activation during treatment, which induced apoptosis (46). Notably, PSME3 is also involved in mediating the deleted in breast cancer 1 (DBC1) regulation of DNA damage-induced apoptosis (47). PSME3 is involved in apoptosis and plays important roles through different pathways; however, the potential antiapoptotic mechanism of PSME3 in LUAD needs to be investigated further.
We also found that PSME3 promotes the migration and invasion of LUAD cells, which is closely related to EMT. Research has shown that the upregulation of PSME3 expression in oral submucous fibrosis contributes to EMT in epithelial cells (48). In thyroid cancer, PSME3-deficient cells showed reduced migration and invasion, and upregulated E-cadherin and Smurf2 expression (49). In addition, PSME3 has been shown to induce EMT in breast cancer, which increases cell migration and invasion (37). However, in hepatocellular carcinoma, insulin-like growth factor 1 (IGF-1) most likely promotes hepatocellular carcinoma (HCC) cell growth and metastasis by blocking UPS-mediated cathepsin B (CTSB) degradation, which can be reversed by overexpressing PSME3 (50). In summary, PSME3 has been shown to promote migration and invasion in most diseases; however, in a small number of cases, it has been shown to inhibit migration and invasion. Therefore, the PSME3 promotion of cell migration and invasion by EMT is not suitable for clinical application. The underlying mechanism needs to be further elucidated.
In addition to its ability to promote non-cancer-related pathways, such as endothelial damage in diabetes by inhibiting the HMGA2-GLUT1 pathway (51), accelerating cardiac hypertrophy by decreasing the PP2Acα-SOD2 pathway (52), and affecting the function of asthmatic airway smooth muscle cells through the AMPK/mTOR pathway (53), PSME3 also promotes numerous cancer-related pathways, such as the Wnt/β-catenin pathway in skin cancers and melanoma (43,54), the Smad7-TGF-β pathway in thyroid cancer cells (27), and the nuclear factor kappa B (NF-κB) pathway in multiple myeloma (55). In the present study, we performed transcriptome sequencing, and a KEGG pathway analysis of the transcriptome data revealed that PSME3 knockdown significantly affected the TGF-β pathway. As members of the Smad family are downstream and have been shown to play roles in thyroid cancer, we focused on the TGF-β/SMAD signaling pathway, and the WB results confirmed that PSME3 affected this pathway. However, the direct molecular mechanism by which PSME3 regulates the TGF-β/SMAD pathway needs to be explored further.
The functions of PSME3 extend far beyond those investigated in this study, and include contributing to tumor angiogenesis in oral squamous cell carcinoma (29), controlling Th17 cell differentiation and autoimmune inflammation by regulating dendritic cells (56), and regulating the energetic homeostasis of tumor cells (57). All these results indicate that PSME3 has a wide range of mechanisms of action and has broad research prospects.
Conclusions
In this study, we confirmed that PSME3 was abnormally highly expressed in lung adenocarcinoma and was associated with poor prognosis of patients. PSME3 mediated lung adenocarcinoma progression by promoting proliferation, metastasis, and invasion of lung adenocarcinoma cells, accelerating cell cycle progression, and inhibiting apoptosis. Mechanistically, PSME3 acted through the TGF-β/SMAD signaling pathway. Overall, PSME3 promotes lung adenocarcinoma progression by regulating the TGF-β/SMAD signaling pathway, which provides a potential new therapeutic target for lung adenocarcinoma and offers new options for lung adenocarcinoma treatment, and it is expected to improve the prognosis of patients.
Acknowledgments
We would like to thank the Research Centre for Basic Medical Sciences of Qilu Hospital of Shandong University for providing the experimental platform.
Funding: This study received funding from
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-340/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-340/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-340/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-340/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of Qilu Hospital (No. KYLL-2021[KS]-1053), and informed consent was obtained from each patient. The animal experiments were performed under a project license (No. DWLL-2023-056) granted by the Animal Research Ethics Committee of Qilu Hospital, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang C, Tan S, Li J, et al. CircRNAs in lung cancer - Biogenesis, function and clinical implication. Cancer Lett 2020;492:106-15. [Crossref] [PubMed]
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Lin W, Chen Y, Wu B, et al. Identification of the pyroptosis related prognostic gene signature and the associated regulation axis in lung adenocarcinoma. Cell Death Discov 2021;7:161. [Crossref] [PubMed]
- Fricker LD. Proteasome Inhibitor Drugs. Annu Rev Pharmacol Toxicol 2020;60:457-76. [Crossref] [PubMed]
- Park J, Cho J, Song EJ. Ubiquitin-proteasome system (UPS) as a target for anticancer treatment. Arch Pharm Res 2020;43:1144-61. [Crossref] [PubMed]
- Narayanan S, Cai CY, Assaraf YG, et al. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist Updat 2020;48:100663. [Crossref] [PubMed]
- Lv H, Wei GY, Guo CS, et al. 20S proteasome and glyoxalase 1 activities decrease in erythrocytes derived from Alzheimer's disease patients. Neural Regen Res 2020;15:178-83. [Crossref] [PubMed]
- Le Guerroué F, Youle RJ. Ubiquitin signaling in neurodegenerative diseases: an autophagy and proteasome perspective. Cell Death Differ 2021;28:439-54. [Crossref] [PubMed]
- Qiu M, Chen J, Li X, et al. Intersection of the Ubiquitin-Proteasome System with Oxidative Stress in Cardiovascular Disease. Int J Mol Sci 2022;23:12197. [Crossref] [PubMed]
- Yadav D, Lee JY, Puranik N, et al. Modulating the Ubiquitin-Proteasome System: A Therapeutic Strategy for Autoimmune Diseases. Cells 2022;11:1093. [Crossref] [PubMed]
- Boon L, Belmondo T, Vulsteke JB, et al. Anti-Ki/anti-PA28γ autoantibodies contribute to the HEp-2 indirect immunofluorescence nuclear speckled pattern. Clin Chem Lab Med 2023;61:435-41. [Crossref] [PubMed]
- Mofers A, Pellegrini P, Linder S, et al. Proteasome-associated deubiquitinases and cancer. Cancer Metastasis Rev 2017;36:635-53. [Crossref] [PubMed]
- Zeng G, Yu Q, Zhuang R, et al. Recent advances and future perspectives of noncompetitive proteasome inhibitors. Bioorg Chem 2023;135:106507. [Crossref] [PubMed]
- Walhelm T, Gunnarsson I, Heijke R, et al. Clinical Experience of Proteasome Inhibitor Bortezomib Regarding Efficacy and Safety in Severe Systemic Lupus Erythematosus: A Nationwide Study. Front Immunol 2021;12:756941. [Crossref] [PubMed]
- Gandolfi S, Laubach JP, Hideshima T, et al. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev 2017;36:561-84. [Crossref] [PubMed]
- Raninga PV, Lee A, Sinha D, et al. Marizomib suppresses triple-negative breast cancer via proteasome and oxidative phosphorylation inhibition. Theranostics 2020;10:5259-75. [Crossref] [PubMed]
- Chen X, Barton LF, Chi Y, et al. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell 2007;26:843-52. [Crossref] [PubMed]
- Li X, Amazit L, Long W, et al. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell 2007;26:831-42. [Crossref] [PubMed]
- Li X, Lonard DM, Jung SY, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 2006;124:381-92. [Crossref] [PubMed]
- Zhang Z, Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J 2008;27:852-64. [Crossref] [PubMed]
- Boulpicante M, Darrigrand R, Pierson A, et al. Tumors escape immunosurveillance by overexpressing the proteasome activator PSME3. Oncoimmunology 2020;9:1761205. [Crossref] [PubMed]
- Lee HJ, Alirzayeva H, Koyuncu S, et al. Cold temperature extends longevity and prevents disease-related protein aggregation through PA28γ-induced proteasomes. Nat Aging 2023;3:546-66. [Crossref] [PubMed]
- Jonik-Nowak B, Menneteau T, Fesquet D, et al. PIP30/FAM192A is a novel regulator of the nuclear proteasome activator PA28γ. Proc Natl Acad Sci U S A 2018;115:E6477-86. [Crossref] [PubMed]
- Guo J, Hao J, Jiang H, et al. Proteasome activator subunit 3 promotes pancreatic cancer growth via c-Myc-glycolysis signaling axis. Cancer Lett 2017;386:161-7. [Crossref] [PubMed]
- Yu L, Li JJ, Liang XL, et al. PSME3 Promotes TGFB1 Secretion by Pancreatic Cancer Cells to Induce Pancreatic Stellate Cell Proliferation. J Cancer 2019;10:2128-38. [Crossref] [PubMed]
- Wang Q, Pan F, Li S, et al. The prognostic value of the proteasome activator subunit gene family in skin cutaneous melanoma. J Cancer 2019;10:2205-19. [Crossref] [PubMed]
- Jiao C, Li L, Zhang P, et al. REGγ ablation impedes dedifferentiation of anaplastic thyroid carcinoma and accentuates radio-therapeutic response by regulating the Smad7-TGF-β pathway. Cell Death Differ 2020;27:497-508. [Crossref] [PubMed]
- Wang Y, Xu J, Zhu X, et al. MicroRNA-130a-3p impedes the progression of papillary thyroid carcinoma through downregulation of KPNB1 by targeting PSME3. Endocrine 2023;82:96-107. [Crossref] [PubMed]
- Liu S, Liu D, Zeng X, et al. PA28γ acts as a dual regulator of IL-6 and CCL2 and contributes to tumor angiogenesis in oral squamous cell carcinoma. Cancer Lett 2018;428:192-200. [Crossref] [PubMed]
- Jiang M, Zhu Y, Yu H. Ginsenoside 20(S)-Rg3 suppresses cell viability in esophageal squamous cell carcinoma via modulating miR-324-5p-targeted PSME3. Hum Exp Toxicol 2021;40:1974-84. [Crossref] [PubMed]
- Schizas D, Kapsampelis P, Mylonas KS MD. Adenosquamous Carcinoma of the Esophagus: A Literature Review. J Transl Int Med 2018;6:70-3. [Crossref] [PubMed]
- Chen ZM, Kai Z, Fang J, et al. The prognosis value of proteasome activator subunit 3 expression in gastric cancer. J Physiol Pharmacol 2022; [Crossref] [PubMed]
- Liu C, Yang J, Wu H, et al. Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3. Onco Targets Ther 2019;12:6525-34. [Crossref] [PubMed]
- Song W, Guo C, Chen J, et al. Silencing PSME3 induces colorectal cancer radiosensitivity by downregulating the expression of cyclin B1 and CKD1. Exp Biol Med (Maywood) 2019;244:1409-18. [Crossref] [PubMed]
- Gao Z, Jiang J, Hou L, et al. Dysregulation of MiR-144-5p/RNF187 Axis Contributes To the Progression of Colorectal Cancer. J Transl Int Med 2022;10:65-75. [Crossref] [PubMed]
- Wei X, Sun K, Li S, et al. PSME3 induces radioresistance and enhances aerobic glycolysis in cervical cancer by regulating PARP1. Tissue Cell 2023;83:102151. [Crossref] [PubMed]
- Yi Z, Yang D, Liao X, et al. PSME3 induces epithelial-mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp Cell Res 2017;358:87-93. [Crossref] [PubMed]
- Peng D, Fu M, Wang M, et al. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer 2022;21:104. [Crossref] [PubMed]
- Frayssinhes JA, Cerruti F, Laulin J, et al. PA28γ-20S proteasome is a proteolytic complex committed to degrade unfolded proteins. Cell Mol Life Sci 2021;79:45. [Crossref] [PubMed]
- Thomas TA, Smith DM. Proteasome activator 28γ (PA28γ) allosterically activates trypsin-like proteolysis by binding to the α-ring of the 20S proteasome. J Biol Chem 2022;298:102140. [Crossref] [PubMed]
- Yu Z, Wei X, Liu L, et al. Indirubin-3'-monoxime acts as proteasome inhibitor: Therapeutic application in multiple myeloma. EBioMedicine 2022;78:103950. [Crossref] [PubMed]
- Gupta S, Silveira DA, Mombach JCM. Towards DNA-damage induced autophagy: A Boolean model of p53-induced cell fate mechanisms. DNA Repair (Amst) 2020;96:102971. [Crossref] [PubMed]
- Chen H, Gao X, Sun Z, et al. REGγ accelerates melanoma formation by regulating Wnt/β-catenin signalling pathway. Exp Dermatol 2017;26:1118-24. [Crossref] [PubMed]
- Moncsek A, Gruner M, Meyer H, et al. Evidence for anti-apoptotic roles of proteasome activator 28γ via inhibiting caspase activity. Apoptosis 2015;20:1211-28. [Crossref] [PubMed]
- Shi Y, Luo X, Li P, et al. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett 2015;358:27-36. [Crossref] [PubMed]
- Yang L, Zhou J, Meng F, et al. G1 phase cell cycle arrest in NSCLC in response to LZ-106, an analog of enoxacin, is orchestrated through ROS overproduction in a P53-dependent manner. Carcinogenesis 2019;40:131-44. [Crossref] [PubMed]
- Magni M, Ruscica V, Buscemi G, et al. Chk2 and REGγ-dependent DBC1 regulation in DNA damage induced apoptosis. Nucleic Acids Res 2014;42:13150-60. [Crossref] [PubMed]
- Xie C, Li Z, Hua Y, et al. Identification of a BRAF/PA28γ/MEK1 signaling axis and its role in epithelial-mesenchymal transition in oral submucous fibrosis. Cell Death Dis 2022;13:701. [Crossref] [PubMed]
- Bhatti MZ, Pan L, Wang T, et al. REGγ potentiates TGF-β/Smad signal dependent epithelial-mesenchymal transition in thyroid cancer cells. Cell Signal 2019;64:109412. [Crossref] [PubMed]
- Lei T, Ling X. IGF-1 promotes the growth and metastasis of hepatocellular carcinoma via the inhibition of proteasome-mediated cathepsin B degradation. World J Gastroenterol 2015;21:10137-49. [Crossref] [PubMed]
- Xie Y, Gao R, Gao Y, et al. The proteasome activator REGγ promotes diabetic endothelial impairment by inhibiting HMGA2-GLUT1 pathway. Transl Res 2022;246:33-48. [Crossref] [PubMed]
- Xie Y, Gao Y, Gao R, et al. The proteasome activator REGγ accelerates cardiac hypertrophy by declining PP2Acα-SOD2 pathway. Cell Death Differ 2020;27:2952-72. [Crossref] [PubMed]
- Xiao Y, Zhu H, Lei J, et al. MiR-182/Sestrin2 affects the function of asthmatic airway smooth muscle cells by the AMPK/mTOR pathway. J Transl Int Med 2023;11:282-93. [Crossref] [PubMed]
- Li L, Dang Y, Zhang J, et al. REGγ is critical for skin carcinogenesis by modulating the Wnt/β-catenin pathway. Nat Commun 2015;6:6875. [Crossref] [PubMed]
- Liu S, Zheng LL, Zhu YM, et al. Knockdown of REGγ inhibits the proliferation and migration and promotes the apoptosis of multiple myeloma cells by downregulating NF-κB signal pathway. Hematology 2018;23:277-83. [Crossref] [PubMed]
- Zhou L, Yao L, Zhang Q, et al. REGγ controls Th17 cell differentiation and autoimmune inflammation by regulating dendritic cells. Cell Mol Immunol 2020;17:1136-47. [Crossref] [PubMed]
- Sun L, Fan G, Shan P, et al. Regulation of energy homeostasis by the ubiquitin-independent REGγ proteasome. Nat Commun 2016;7:12497. [Crossref] [PubMed]


