
Correlation analysis between driver gene mutation and clinicopathological features in lung adenocarcinoma based on real-world cumulative clinical data
Highlight box
Key findings
• The study identified driver gene mutations in a substantial 89.09% of the 440 lung adenocarcinoma (LUAD) patients.
• Epidermal growth factor receptor (EGFR) mutations exhibited varied distribution, with the majority in the protein tyrosine kinase catalytic domain. Single gene locus mutations occurred in 343 patients, covering all tested genes.
• EGFR mutations were more prevalent in non-smoking and female patients, while KRAS mutations were associated with male patients and smokers. ROS1 mutations correlated with larger tumor diameters, and RET mutations were more common in smokers.
What is known and what is new?
• Prior knowledge includes the prevalence of EGFR, KRAS, and other driving mutations in LUAD. The association of EGFR mutations with non-smoking and female patients, and KRAS mutations with smokers, aligns with existing understanding.
• Identifying a broad spectrum of mutations can help clinicians tailor treatments to individual patients, particularly for those with rare or co-occurring mutations. The association of ROS1 mutations with larger tumor diameters and RET mutations with smoking represents novel insights into the clinicopathological features associated with specific mutations.
What is the implication, and what should change now?
• Understanding the diverse genetic mutations in LUAD patients is crucial for personalized treatment strategies. The association of mutations with clinical features suggests potential biomarkers for prognosis and treatment response.
• Next-generation sequencing may be beneficial for clinicians to identify a broader spectrum of mutations for LUAD patients.
Introduction
Approximately 80% of lung cancer cases are classified as non-small cell lung cancer (NSCLC). The most common subtype of NSCLC is lung adenocarcinoma (LUAD), which accounts for approximately 55% of NSCLC (1). LUAD has a higher incidence and case fatality rate compared to other types of malignant tumors and exhibits a high level of genetic heterogeneity (2,3). Despite significant improvements in clinical treatment through traditional surgery and the combination of radiotherapy and chemotherapy, overall efficacy and prognosis remain limited. In the past 20 years, with the development of tumor molecular biology and the gradual deepening of gene research, the diagnosis and treatment of LUAD have undergone significant changes (4-7). The development and application of targeted drugs have extended survival and improved LUAD patients’ quality of life (8). The carcinogenic mutation of the epidermal growth factor receptor (EGFR) tyrosine kinase domain showed a significant clinical response to the first-generation EGFR tyrosine kinase inhibitor (TKI) gefitinib in 2004 (9), ushering in the era of targeted treatment for NSCLC. Three years later, the discovery of ALK rearrangement broadened the scope of targeted genomic changes in the disease (10). Subsequently, some other driver events with strong transformation potential were reported, including somatic oncogenic mutations of ROS1, RET, BRAF, gene insertion of HER2, and exon 14 skipping of MET proto-oncogene (8,11). Driver genes are essential predictors of targeted therapeutic efficacy. Detecting driver gene mutations in LUAD patients can help to screen for targeted drugs and improve patient survival benefits (12-15). This study analyzed the mutations of driver genes (EGFR, KRAS, ALK, ROS1, RET, MET, BRAF, HER2, PIK3CA and NRAS) in LUAD and their correlation with clinical pathological features, providing a theoretical basis for promoting multi-gene joint detection. We present this article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-409/rc).
Methods
Patients and tumor samples
In this retrospective study, a total of 440 LUAD patients who met the inclusion criteria were selected from Sir Run Run Shaw Hospital. The inclusion criteria were: (I) the result of cytological or pathological diagnosis was LUAD; (II) patient was not receiving any radiotherapy/chemotherapy treatment before the operation; (III) traceability of the multi-gene joint test result could be implemented. Patients were excluded based on the following criteria: (I) patients without clinical pathological information; (II) gene testing experiment failed. All surgical lung tissue specimens were classified by two independent pathologists based on histological results. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Ethics Committee of Sir Run Run Shaw Hospital (No. 20230488). Informed consent has been taken from the participants before taking part.
Sample preparation and DNA extraction
LUAD tissue samples, with a tumor content exceeding 20%, were collected during surgical procedures. The QIAamp DNA FFPE Tissue Kit (Qiagen, Dusseldorf, Germany) was used to extract these samples, adhering to the manufacturer’s guidelines. DNA concentration was evaluated using Qubit 3.0 (Thermo Fisher Scientific, Waltham, USA), while DNA size distribution was analyzed with the Qsep100 system (Bioptic, Taiwan, China). These standardized protocols were used to ensure accurate and reproducible results.
Next-generation sequencing (NGS) library preparation
This study utilized the CleanPlexTM sequencing kit (Paragon Genomics, Silicon Valley, USA) following the optimized protocol provided by the manufacturer to generate sequencing libraries. The current study employed multiplex PCR to target 10 LUAD-associated genes and enrich 200 g of genomic DNA. Beckman Coulter’s Agencourt AMPure XP beads were used to purify the DNA library. The NGS library was further purified and quantified using the Qubit 1X dsDNA Assay Kit from Thermo Fisher Scientific. Fragment size distribution was analyzed using Bioptic’s Qsep100 in Taiwan, China. Genetic testing was conducted in a laboratory certified by the College of American Pathologists (CAP) to ensure that high quality standards were met. A panel of 10 LUAD-associated genes was employed to identify single nucleotide variants, deletions, insertions, delins mutations, duplications, and fusion.
Sequencing and bioinformatics analysis
Library sequencing was performed on the Illumina NextSeq 500 platform (Illumina, San Diego, USA), achieving an average depth of over 1,000×. Variants with a frequency of at least 1% were designated as mutations. Paired-end sequencing data from libraries in FASTQ format were aligned to the human genome (hg19) using Burrows-Wheeler Aligner (BWA-MEM). To identify potential fusion detection points, the coverage depth of adjacent sites and fusion breakpoints was calculated. ANNOVAR21, along with the hg19 reference genome, standard databases, and functional prediction programs, were used to perform SNV and indel annotation.
Statistical analysis
All analyses in this academic paper were conducted via R version 4.1.1 (2021-08-10). Continuous variables were presented using mean and standard deviation or median and interquartile ranges, and comparisons were made using the Student’s t-test or the Mann-Whitney U test. Subgroup analyses were conducted using the Chi-square test, with Fisher’s exact test applied for small sample sizes. Kendall’s tau coefficient was employed for correlation analyses. All analyses conducted in this paper were two-tailed, and a significance level of P≤0.05 was used as the standard criterion for determining statistical significance. This paper addresses an audience of our peers and esteemed instructors.
Results
Clinicopathological characteristics of 440 LUAD patients
Four hundred and forty LUAD patients were enrolled in this study. In this study, there was no gender bias at the onset of LUAD (available online: https://cdn.amegroups.cn/static/public/tlcr-24-409-1.xlsx). There were 213 (48.41%) male patients and 227 (51.59%) female patients with a male-to-female ratio of 1:1.0657. The age range of the LUAD patients was 26 to 89 years, with a median age of 65 years. More than 60% of all patients with LUAD were elderly aged >60 years old. Three-quarters of LUAD occurred in patients without a history of smoking. A minority of LUAD patients showed multiple pulmonary lesions, while the majority (over 80%) exhibited singular lesions. Sixty percent of the patients were diagnosed with stage I LUAD. The main patient groups of LUAD were tumor diameter ≤3 cm or without any lymph node metastasis or any distant metastasis. The incidence of the right lung was 1.2 times higher than that of the left lung. The upper lobe incidence rate was 1.5 times higher than that of the lower lobe (Table 1).
Table 1
| Characteristics | No. of patients | % |
|---|---|---|
| Gender | ||
| Male | 213 | 48.41 |
| Female | 227 | 51.59 |
| Age, years | ||
| ≤60 | 165 | 37.50 |
| >60 | 275 | 62.50 |
| Smoke | ||
| Yes | 36 | 8.18 |
| No | 333 | 75.68 |
| Quit | 56 | 12.73 |
| Unknown | 15 | 3.41 |
| Lesions | ||
| Single | 366 | 83.18 |
| Multiple | 74 | 16.82 |
| Diameter | ||
| ≤3 | 358 | 81.36 |
| >3 and ≤5 | 62 | 14.09 |
| >5 | 20 | 4.55 |
| Stage | ||
| Stage I | 264 | 60.00 |
| Stage II | 19 | 4.32 |
| Stage III | 56 | 12.73 |
| Stage IV | 41 | 9.32 |
| Unknown | 60 | 13.64 |
| Distant metastasis | ||
| None | 349 | 79.32 |
| Yes | 91 | 20.68 |
| Lymph node metastasis | ||
| Yes | 85 | 19.32 |
| No | 355 | 80.68 |
| Location of lesions | ||
| Left | 196 | 44.55 |
| Right | 237 | 53.86 |
| Left & right | 5 | 1.14 |
| Others | 2 | 0.45 |
| Location of lesions (pulmonary lobe) | ||
| Upper lobe | 223 | 50.68 |
| Lower lobe | 146 | 33.18 |
| Upper & lower lobe | 38 | 8.64 |
| Others | 33 | 7.50 |
Driver gene mutation characteristics
Among the 440 LUAD patients, 48 did not have detectable driver gene mutations (available online: https://cdn.amegroups.cn/static/public/tlcr-24-409-1.xlsx). The percentage of patients with a driver gene mutation was 89.09%. 392 LUAD patients carried at least one driver gene mutation, including 343 patients with a single gene mutation and 49 patients with two or more driver gene mutations. The results of the multi-gene analysis show that EGFR (299, 67.95%) had the highest mutation rate among 10 genes, followed by KRAS (33, 7.50%), MET (26, 5.91%), HER2 (24, 5.45%), ALK (17, 3.86%), RET (14, 3.18%), ROS1 (10, 2.27%), BRAF (10, 2.27%), PIK3CA (8, 1.82%) and NRAS (1, 0.23%) (Figure 1).
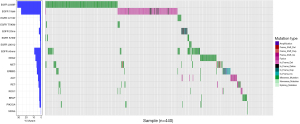
Among the 299 patients with EGFR mutations, 260 patients carried only one EGFR mutation site, 38 patients carried two EGFR mutation sites, and only one patient carried three EGFR mutation sites simultaneously. The most common EGFR mutations were L858R and exon19 deletion, which accounted for 46.82% and 39.13% of patients with EGFR mutations respectively. The EGFR T790M mutation was found in six TKI-naive LUAD patients. Three rare EGFR mutations, G719X (7, 2.34%), S768I (6, 2.01%), and L861X (6, 2.01%) were identified. We detected 69 different EGFR mutation sites in the 299 patients with EGFR mutations which were distributed in the protein tyrosine kinase catalytic domain (56, 81.16%), Furin-like cysteine-rich region (9, 13.04%), receptor binding domain (3, 4.35%), and EGFR transmembrane domain (1, 1.45%) (Figure 2). Forty-three patients with EGFR mutations also carried other driver mutations (Table 2). The gene with the highest incidence of EGFR co-mutation was MET [14], while other genes were ERBB2 [7], RET [6], PIK3CA [5], ROS1 [5], ALK [3] and KRAS [1].
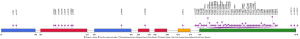
Table 2
| Co-mutated gene | No. of patients | % of EGFR mutation (n=299) | % of LUAD (n=440) |
|---|---|---|---|
| EGFR + MET | 14 | 4.68 | 3.18 |
| EGFR + ERBB2 | 7 | 2.34 | 1.59 |
| EGFR + RET | 6 | 2.01 | 1.36 |
| EGFR + PIK3CA | 5 | 1.67 | 1.14 |
| EGFR + ROS1 | 5 | 1.67 | 1.14 |
| EGFR + ALK | 3 | 1.00 | 0.68 |
| EGFR + KRAS | 1 | 0.33 | 0.23 |
| EGFR + ROS1 + RET | 1 | 0.33 | 0.23 |
| EGFR + MET + RET | 1 | 0.33 | 0.23 |
EGFR, epidermal growth factor receptor; LUAD, lung adenocarcinoma.
We also observed 23 patients with fusion mutations. ALK fusion occurred in 15 patients, with a total of 5 companion genes, the most common of which was EML4 in 12 cases, and other rare companion genes were DCTN1, AC011239.1, STRN and FAM179A (Table S1). There were three subtypes of EML4-ALK fusion, including three cases of E6:A20, five cases of E13:A20 and one case of E18:A20. RET fusion occurred in four patients, and the partner genes were KIF13A, KIF5B, KCND2 and LDLR. ROS1 fusion mutation was detected in four patients, with one patient fusing with two partner genes, STK39 and EZR, and other partner genes being FAM65B, PKHD1 and GLI3.
Associations between driver gene mutation and clinicopathological features in LUAD
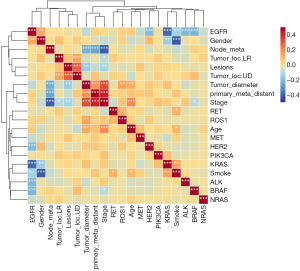
In this study, driver gene mutations were more likely to occur in female LUAD patients (P=0.03). EGFR mutations also tended to occur in female patients (P=0.0036). There were differences in driving gene mutations among LUAD patients with different smoking histories (P=0.057), but they were not significant. However, EGFR mutations were prevalent in LUAD patients without a history of smoking (P=0.0005). No significant differences were detected in driver gene or EGFR mutations concerning age, lesion placement, lesion quantity, tumor size, presence of lymph node metastasis, lesion metastasis, and clinical stage (Table 3). The Kendall correlation significance test indicated that EGFR mutations were mutually exclusive with mutations in KRAS, ALK, ERBB2, and BRAF (P<0.001) (refer to Figure 3). EGFR mutations were predominantly observed in women and non-smokers (P<0.01), whereas KRAS mutations were more prevalent among male smokers (P<0.01). ROS1 mutations and RET mutations were prone to occur simultaneously (P<0.05). Patients with ROS1 mutations had larger tumor diameters (P<0.01). RET mutations were more prevalent in individuals who smoke (P<0.05).
Table 3
| Characteristics | Gene mutation | EGFR mutation | |||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | P value | No | Yes | P value | ||
| Gender | 0.03 | 0.0036 | |||||
| Male | 31 | 182 | 83 | 130 | |||
| Female | 17 | 210 | 58 | 169 | |||
| Age, years | 0.43 | 0.33 | |||||
| ≤60 | 21 | 144 | 58 | 107 | |||
| >60 | 27 | 248 | 83 | 192 | |||
| Smoking history | 0.057 | 0.0005 | |||||
| No | 30 | 303 | 93 | 240 | |||
| Yes | 4 | 32 | 16 | 20 | |||
| Quit | 11 | 45 | 29 | 27 | |||
| Location of lesions (pulmonary lobe) | 0.44 | 0.31 | |||||
| Upper lobe | 27 | 196 | 67 | 156 | |||
| Lower lobe | 12 | 134 | 55 | 91 | |||
| Upper & lower lobe | 5 | 33 | 12 | 26 | |||
| Location of lesions | 0.81 | 0.93 | |||||
| Left | 21 | 175 | 63 | 133 | |||
| Right | 26 | 211 | 76 | 161 | |||
| Left & right | 1 | 4 | 2 | 3 | |||
| No. of primary focus | 0.87 | 0.84 | |||||
| Single lesion | 39 | 326 | 116 | 249 | |||
| Multiple lesions | 9 | 65 | 25 | 49 | |||
| Tumor diameter, cm | 0.12 | 0.056 | |||||
| ≤3 | 37 | 321 | 108 | 250 | |||
| >3 and ≤5 | 6 | 56 | 22 | 40 | |||
| >5 | 5 | 15 | 11 | 9 | |||
| Lymphatic metastasis | 0.49 | 0.95 | |||||
| Yes | 7 | 78 | 27 | 58 | |||
| No | 41 | 314 | 114 | 241 | |||
| Tumor metastasis | 0.59 | 0.50 | |||||
| Yes | 8 | 83 | 26 | 65 | |||
| No | 40 | 309 | 115 | 234 | |||
| Tumor stage | 0.42 | 0.65 | |||||
| Stage I | 31 | 233 | 87 | 177 | |||
| Stage II | 0 | 19 | 4 | 15 | |||
| Stage III | 5 | 51 | 20 | 36 | |||
| Stage IV | 5 | 36 | 12 | 29 | |||
EGFR, epidermal growth factor receptor.
Discussion
Mutations in driver genes play a crucial role in determining the response to targeted therapies in LUAD patients. Targeted therapy guided by gene mutations significantly improves treatment choices and survival benefits for LUAD patients (11). In addition to commonly activating EGFR mutations, targets such as ALK rearrangement, ROS1 rearrangement, RET rearrangement, BRAF V600E, MET exon 14 skipping mutation, and KRAS G12C have gradually been approved for targeted therapy (8). The number of gene mutations associated with targeted therapy has increased from one gene to several genes (16,17). Therefore, a comprehensive examination of genetic mutations in patients is essential. Multi-gene targeted sequencing is a rapid and cost-effective detection method that can provide comprehensive recommendations for LUAD-targeted drugs. In this analysis, we employed a multi-gene targeted sequencing technology to comprehensively evaluate driver gene mutations in 440 LUAD patients. The top three driver genetic mutations were EGFR, KRAS, and MET. Typically, EGFR and KRAS mutations do not occur together, but we observed a patient with concurrent EGFR and KRAS mutations, suggesting primary resistance to EGFR inhibitors. In addition, we identified five patients with EGFR L858R mutations or exon 19 deletions accompanied by EGFR T790M mutations, indicating resistance to the first and second-generation EGFR-TKIs. These findings suggest that clinicians need to carefully select EGFR-TKIs targeted therapy, and consider the patient’s benefits when developing treatment plans. Furthermore, we found that 27 patients detected only one KRAS hotspot mutation, G12X or Q61H. Nine patients carried BRAF mutations, six patients had MET mutations, 12 patients developed HER2 insertion mutations, and 17 patients formed gene fusions that included 12 ALK fusions, three RET fusions, and two ROS1 fusions. It is important to note that these patients carried only one gene mutation. There was a rare case of ALK fusion, DCTN1/ALK (D24: A20). Gao et al. (18) reported one case of DCTN1/ALK fusion that achieved partial response after receiving cabozantinib therapy. Adding to the complexity of genomic landscapes in LUAD, we report a patient with a novel fusion involving STK39 and EZR, where EZR’s coiled-coil domain contrasts with the absence of such a structure in STK39. This discrepancy underscores the need for further exploration of STK39’s role in this fusion context and highlights the importance of investigating these gene fusions as potential driver mutations. It is necessary to detect multiple gene mutations in LUAD patients to increase the chances of obtaining potentially beneficial information about target genes and to reduce the likelihood of negative results from single-gene testing. Previous studies have shown that LUAD’s molecular characteristics are influenced by factors such as the environment, family and lifestyle (19,20). Some studies emphasized the close relationship between environmental factors, such as gender and smoking, and LUAD molecular characteristics (21-23). Wu et al. (24) analyzed 506 NSCLC patients, and the results showed that the EGFR mutation rate in non-smoking patients was higher than that in smoking patients and mutation rate was higher in female patients than male patients. An analysis by An et al. (25) on 524 NSCLC patients revealed that the gene most frequently altered in non-smoking adenocarcinoma patients was EGFR. Our analysis revealed a notable correlation between gene mutations and clinicopathological factors including gender, smoking history, and lesion site. Specifically, EGFR mutations were significantly more frequent among non-smokers and female patients (P<0.01). However, our analysis indicated that KRAS mutations were more frequent in male patients and smokers (P<0.01). The observation that ROS1 mutations are associated with larger tumor diameters may be due to the aggressive nature of ROS1-driven tumors, which activate pathways that promote cell proliferation and survival, leading to increased tumor growth. Studies have shown that ROS1 rearrangements can activate downstream signaling pathways, such as PI3K/AKT and MAPK/ERK, which are crucial for cell proliferation and survival (26,27). It is plausible that similar mechanisms could be involved in tumors with ROS1 mutations, although further research is needed to elucidate this association. Interestingly, while our study found a higher prevalence of RET mutations in smokers, previous literature indicates that RET fusions occur more frequently in non-smokers (28). This discrepancy suggests different underlying mechanisms for RET mutations and fusions. Smoking-related carcinogens are known to increase the overall mutational burden, potentially leading to RET mutations through DNA-damaging effects. However, RET fusions may not be as strongly influenced by smoking status. Future studies are necessary to clarify these associations and to understand the specific effects of smoking on RET mutations and fusions.
Targeted therapies have significantly advanced the treatment of LUAD by focusing on specific genetic alterations (7,8). EGFR mutations are effectively targeted by drugs such as gefitinib, erlotinib, and osimertinib. ALK rearrangements respond well to inhibitors like crizotinib, alectinib, and brigatinib. ROS1 rearrangements can be treated with crizotinib and entrectinib. For BRAF V600E mutations, the combination of dabrafenib and trametinib is effective. Additionally, MET amplifications and METex14 skipping mutations can be targeted by drugs like capmatinib and tepotinib. These therapies offer significant improvements in patient outcomes and quality of life.
Meanwhile, identifying a broad spectrum of mutations can help clinicians tailor treatments to individual patients, particularly for those with rare or co-occurring mutations, as seen in Table 2. In LUAD, EGFR mutations may co-occur with other genetic alterations, impacting disease progression, treatment response, and prognosis. Notably as reports, TP53 mutations are commonly found alongside EGFR mutations, often indicating a poorer prognosis and potentially influencing the response to EGFR inhibitors (29). PIK3CA mutations can co-exist with EGFR mutations, possibly affecting the efficacy of EGFR-targeted therapies and contributing to disease progression (30). Although rare, ALK rearrangements can also occur alongside EGFR mutations, typically resulting in reduced efficacy of single-agent targeted therapies and necessitating combined treatment approaches. While KRAS mutations are generally considered mutually exclusive with EGFR mutations, their occasional co-occurrence indicates complex tumor biology and typically poorer prognosis. MET amplification is a known mechanism of acquired resistance to EGFR inhibitors, requiring alternative therapies such as MET inhibitors (31). HER2, BRAF mutations and ROS1 rearrangements, though less common, also represent significant challenges, often indicating resistance to standard EGFR-targeted treatments. There are also reported cases of triple alterations involving EGFR, ROS1, RET, KRAS and ALK mutations, further complicating treatment strategies and necessitating highly personalized therapeutic approaches (32-34). Understanding these co-occurring genetic alterations is crucial for developing personalized treatment strategies and improving clinical outcomes for LUAD patients.
This study has several limitations. Firstly, the targeted sequencing panels used had limited capacity, potentially overlooking certain mutations in LUAD patients. We have recognized the importance of NTRK gene fusions and their potential impact on patient management. NTRK gene fusions are recognized as significant oncogenic drivers in various cancers, including LUAD. These fusions involve the NTRK1, NTRK2, and NTRK3 genes, leading to the constitutive activation of the TRK signaling pathway, which promotes tumorigenesis (35). The inclusion of NTRK gene fusions in comprehensive genomic profiling is therefore essential for identifying patients who may benefit from TRK inhibitor therapies. Future studies will include NTRK gene fusions to provide a more complete landscape of actionable genetic alterations in LUAD, ensuring that all potential therapeutic targets are considered. Whole exome sequencing may be a recommended method for a comprehensive understanding of gene alterations. While our study provides valuable insights into the molecular mutation characteristics of LUAD patients and their correlation with clinicopathological features, our study lacked follow-up information on the patients, thus preventing us from analyzing the relationship between molecular mutation characteristics and prognosis. Further research is needed to elucidate the relationship between co-occurring genetic mutations and their impact on LUAD prognosis and treatment outcomes. Collective insights from these investigations will contribute to the development of more effective targeted therapy strategies for LUAD patients. Understanding the prognostic implications of these mutations is crucial for guiding treatment decisions and predicting patient outcomes. Additionally, the widespread implementation of CT lung cancer screening in China has led to the early detection and treatment of an increasing number of early-stage lung cancer cases. This screening practice contributes significantly to the observed predominance of early-stage LUAD in our study. Similarly, the higher age range for EGFR mutations and the predominance of stage I cancers in our cohort reflect this selection bias. Most NSCLC cases present at a later stage (stage III or IV) and are not suitable for surgical resection. For advanced-stage NSCLC patients, we typically perform pathological biopsy to seek targeted therapy for optimal benefit. our study was limited to a single-center retrospective analysis, which may introduce selection bias and limit the generalizability of our findings to broader populations.
Conclusions
In conclusion, our study found that LUAD patients in large centre in China exhibited diverse genetic mutations, which may co-occur simultaneously. Integrated analysis of multiple mutations is essential for accurate diagnosis and effective treatment of this disease. The use of NGS can significantly expand our understanding of gene mutations and facilitate integrated analysis of multiple gene mutations, providing critical evidence for targeted treatment methods. Despite the limitations of this study, these findings hold promise for improving clinical decision-making for LUAD management in patients. Further research is needed to elucidate the relationship between co-occurring genetic mutations and their impact on LUAD prognosis and treatment outcomes. Collective insights from these investigations will contribute to the development of more effective targeted therapy strategies for LUAD patients.
Acknowledgments
The authors express gratitude to all staff members of the Pathology Department at Sir Run Run Shaw Hospital for their invaluable assistance in managing case information. Additionally, special thanks are extended to Tingting Pan from the Precision Diagnosis Center for her coordination of the sequencing data. The authors also acknowledge the editor and reviewers for their constructive feedback, which enhanced the quality of this paper.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-409/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-409/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-409/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-409/coif). S.A. receives honoraria for lectures from MSD, AstraZeneca, Roche, BMS, Merck Serono, Takeda, and support for attending meetings from AstraZeneca, Roche, MSD, Pfizer; and serves on Advisory Board of Roche, BeiGene, Amgen. N.Y., X.Z. and D.S. provided relevant assistance in NGS detection and serve on Dian Diagnostics Group Co., Ltd., Hangzhou, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Sir Run Run Shaw Hospital (No. 20230488). Informed consent has been taken from the participants before taking part.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol 2023;20:624-39. [Crossref] [PubMed]
- Harada G, Yang SR, Cocco E, et al. Rare molecular subtypes of lung cancer. Nat Rev Clin Oncol 2023;20:229-49. [Crossref] [PubMed]
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [Crossref] [PubMed]
- Chaft JE, Rimner A, Weder W, et al. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat Rev Clin Oncol 2021;18:547-57. [Crossref] [PubMed]
- Lengel HB, Mastrogiacomo B, Connolly JG, et al. Genomic mapping of metastatic organotropism in lung adenocarcinoma. Cancer Cell 2023;41:970-985.e3. [Crossref] [PubMed]
- Asahina H, Yamazaki K, Kinoshita I, et al. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer 2006;95:998-1004. [Crossref] [PubMed]
- Król K, Mazur A, Stachyra-Strawa P, et al. Non-Small Cell Lung Cancer Treatment with Molecularly Targeted Therapy and Concurrent Radiotherapy-A Review. Int J Mol Sci 2023;24:5858. [Crossref] [PubMed]
- Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med 2021;27:1345-56. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Greenberg PL, Stone RM, Al-Kali A, et al. NCCN Guidelines® Insights: Myelodysplastic Syndromes, Version 3.2022. J Natl Compr Canc Netw 2022;20:106-17. [Crossref] [PubMed]
- Wolf J, Seto T, Han JY, et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:944-57. [Crossref] [PubMed]
- Solomon BJ, Bauer TM, Mok TSK, et al. Efficacy and safety of first-line lorlatinib versus crizotinib in patients with advanced, ALK-positive non-small-cell lung cancer: updated analysis of data from the phase 3, randomised, open-label CROWN study. Lancet Respir Med 2023;11:354-66. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Chiari R, et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: a multicentre, open-label, single-arm, phase 1-2 trial. Lancet Oncol 2019;20:1691-701. [Crossref] [PubMed]
- Riely GJ, Smit EF, Ahn MJ, et al. Phase II, Open-Label Study of Encorafenib Plus Binimetinib in Patients With BRAFV600-Mutant Metastatic Non-Small-Cell Lung Cancer. J Clin Oncol 2023;41:3700-11. Erratum in: J Clin Oncol 2024;42:245. [Crossref] [PubMed]
- Chang YS, Tu SJ, Chen YC, et al. Mutation profile of non-small cell lung cancer revealed by next generation sequencing. Respir Res 2021;22:3. [Crossref] [PubMed]
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Gao F, Gao F, Wu H, et al. Response to ALK-TKIs in a lung adenocarcinoma patient harboring dual DCTN1-ALK and ALK-CLIP4 rearrangements. Thorac Cancer 2022;13:1088-90. [Crossref] [PubMed]
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. [Crossref] [PubMed]
- Sacher AG, Dahlberg SE, Heng J, et al. Association Between Younger Age and Targetable Genomic Alterations and Prognosis in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:313-20. [Crossref] [PubMed]
- Vineis P, Airoldi L, Veglia F, et al. Environmental tobacco smoke and risk of respiratory cancer and chronic obstructive pulmonary disease in former smokers and never smokers in the EPIC prospective study. BMJ 2005;330:277. [Crossref] [PubMed]
- Kligerman S, White C. Epidemiology of lung cancer in women: risk factors, survival, and screening. AJR Am J Roentgenol 2011;196:287-95. [Crossref] [PubMed]
- Yano T, Haro A, Shikada Y, et al. Non-small cell lung cancer in never smokers as a representative 'non-smoking-associated lung cancer': epidemiology and clinical features. Int J Clin Oncol 2011;16:287-93. [Crossref] [PubMed]
- Wu YL, Zhong WZ, Li LY, et al. Epidermal growth factor receptor mutations and their correlation with gefitinib therapy in patients with non-small cell lung cancer: a meta-analysis based on updated individual patient data from six medical centers in mainland China. J Thorac Oncol 2007;2:430-9. [Crossref] [PubMed]
- An SJ, Chen ZH, Su J, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One 2012;7:e40109. [Crossref] [PubMed]
- Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res 2013;19:4040-5. [Crossref] [PubMed]
- Roskoski R Jr. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol Res 2017;121:202-12. [Crossref] [PubMed]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Kim Y, Lee B, Shim JH, et al. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J Thorac Oncol 2019;14:193-202. [Crossref] [PubMed]
- Wu SG, Chang YL, Yu CJ, et al. The Role of PIK3CA Mutations among Lung Adenocarcinoma Patients with Primary and Acquired Resistance to EGFR Tyrosine Kinase Inhibition. Sci Rep 2016;6:35249. [Crossref] [PubMed]
- Mi J, Huang Z, Zhang R, et al. Molecular characterization and clinical outcomes in EGFR-mutant de novo MET-overexpressed advanced non-small-cell lung cancer. ESMO Open 2022;7:100347. [Crossref] [PubMed]
- Mao Y, Wu S. ALK and ROS1 concurrent with EGFR mutation in patients with lung adenocarcinoma. Onco Targets Ther. 2017;10:3399-3404. [Crossref] [PubMed]
- Tang Z, Zhang J, Lu X, et al. Coexistent genetic alterations involving ALK, RET, ROS1 or MET in 15 cases of lung adenocarcinoma. Mod Pathol 2018;31:307-12. [Crossref] [PubMed]
- Ju L, Han M, Zhao C, et al. EGFR, KRAS and ROS1 variants coexist in a lung adenocarcinoma patient. Lung Cancer 2016;95:94-7. [Crossref] [PubMed]
- Haratake N, Seto T. NTRK Fusion-positive Non-small-cell Lung Cancer: The Diagnosis and Targeted Therapy. Clin Lung Cancer 2021;22:1-5. [Crossref] [PubMed]


