
miR-16-5p specifically inhibits IFN-γ-regulated memory T helper cell differentiation in malignant pleural effusion of non-small cell lung cancer
Highlight box
Key findings
• To the best of our knowledge, for the first time in this study, CD4+CD69+ T cells have been defined as memory CD4+ T cells in malignant pleural effusion (MPE) of non-small cell lung cancer (NSCLC). Similarly, we have also found that miR-16-5p may regulate the decrease of differentiation of naïve CD4+ T cells into memory CD4+CD69+ T cells through its target gene interferon-gamma (IFN-γ) in MPE.
What is known and what is new?
• MicroRNAs that regulate gene expression play a crucial role in the regulating the differentiation of memory Th cells.
• We defined CD4+CD69+ T cells as memory CD4+ T cells in MPE of NSCLC and found that miR-16-5p inhibits IFN-γ-regulated memory Th cell differentiation in MPE of NSCLC.
What is the implication, and what should change now?
• The data presented in this study imply that miR-16-5p plays a significant role in the regulation of the immune response against NSCLC. MiR-16-5p could serve as a future target for the development of novel therapeutic strategies.
Introduction
Malignant pleural effusion (MPE) of non-small cell lung cancer (NSCLC) is a common event that often leads to poor patient prognosis (1-4). The evaluation of the tumor immune microenvironment of MPE is important for the cure of NSCLC because of the development of immune therapy (5). CD4+ T cells is dominant in MPE, including various kinds of cell population and differentiation (6). Abnormal activation of the interferon-gamma (IFN-γ) signaling pathway in MPE of NSCLC disrupts memory T helper (Th) cell differentiation, impairing the anti-tumor immune response. Recent studies have shown that microRNAs (miRNAs), a class of small non-coding RNAs that regulate gene expression, play a crucial role in the regulation of memory Th cell differentiation (7-10). However, whether miRNA can inhibit the differentiation of memory Th cells in MPE of NSCLC has not been reported. In this research, we discuss the current understanding of the role of miR-16-5p in the regulation of memory Th cell differentiation and its therapeutic potential in the management of MPE in NSCLC.
MiRNAs are potent regulators of gene expression that bind to the 3'-untranslated region (UTR) or 5'-UTR of target messenger RNAs (mRNAs), leading to inhibition of mRNA translation or degradation (11-14). Recent studies have shown that aberrant expression of miRNAs plays a crucial role in the pathogenesis of NSCLC, including MPE (15-18).
Th cells play an essential role in the immune response against cancer, and memory Th cells are a subset that provides long-lasting immune protection (2,19-21). A study has shown that Th cells are migrated and maintained in chronic inflammatory lesions through the CD69-myosin light chain 9 (Myl9) system model (22). Myl9 molecule in small vessels of inflamed lungs may play a key role in the migration of activated T cells into inflammatory lesions. Myl9 can activate CD69-expressing T cells to migrate to inflamed lungs. The CD69-Myl9 system may promote the efficient migration of activated CD69-expressing T cells into the inflammatory airway. Therefore, CD69 and Myl9 are required for inflammatory disease induction (22,23). Similarly, another report showed that circulating CCR5− resting memory CD8 T cells promote CCR5 expression in response to environmental inflammatory stimulation, which is critical for recruitment of these cells into lung tissue (24).
Other studies have shown that levels of CD4+ subsets in the blood of NSCLC patients are relatively normal, but CD8+ effector T cells are increased and CD8+ effector memory cells are decreased compared with healthy donors. At the same time, both CD4+ and CD8+ naïve cells in NSCLC patients significantly reduced IFN-γ and tumor necrosis factor-α (TNF-α) production. And NSCLC patients had fewer CD8+ effector cells that produced IFN-γ and TNF-α compared to healthy subjects. In addition, similar results in the production of IFN-γ, TNF-α, and interleukin-17 (IL-17) by CD4+ or CD8+ memory cells were observed in NSCLC patients. The result suggests impaired or dysregulated function of CD4+ and CD8+ T lymphocytes in patients with NSCLC (25). However, in MPE of NSCLC, the differentiation and function of memory Th cells are disrupted, resulting in tumor evasion of immune response. Abnormal activation of the IFN-γ signaling pathway in MPE of NSCLC upregulates the expression of transcription factor T-bet, leading to the differentiation of memory Th cells into IFN-γ producing cells and preventing their differentiation into anti-tumor Th1 or Th17 cells (26-28).
Hence, aberrant activation of the IFN-γ signaling pathway in MPE of NSCLC disrupts memory Th cell differentiation and promotes immune evasion by tumor cells. MiR-16-5p acts as a key regulator of memory Th cell differentiation by inhibiting IFN-γ-induced differentiation and promoting anti-tumor Th1 or Th17 differentiation. We present this article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-505/rc).
Methods
Patients
This study was conducted at the Sun Yat-sen University Cancer Center from January 2020 to December 2021, enrolling 30 patients with NSCLC. The MPE was confirmed with cytological or pathological examinations by at least two experienced pathologists. The inclusion criteria were as follows: pleural effusion pathology or pleural biopsy indicated NSCLC; measurable lesions in the lungs; pleural effusion caused by metastatic lung cancer was excluded; no injection of drugs in the chest within the past month; no heart, liver, kidney, and hematopoietic system and other serious diseases, pregnant women and children, accompanied by mental disorders, and no immune system diseases. The exclusion criteria were as follows: pleural effusion caused by tumor metastasis in other sites except NSCLC; pleural effusion caused by leaking fluid such as hypoproteinemia. At the same time, 30 age- and sex-matched patients, who were clinically diagnosed with benign pleural effusion (BPE) of lung disease and had not received any intervention were collected. Their inclusion criteria included were as follows: patients with no history of tumor; repeated tests confirmed that there were no tumor cells in pleural effusion; tuberculous pleuritis: adenosine deaminase (ADA) increased in pleural effusion (>40 U/L), pleural biopsy indicated the presence of mycobacterium tuberculosis or typical epithelioid granuloma, and pleural effusion could be absorbed after anti-tuberculous therapy; parapneumonic effusion: there are acute infection symptoms of pneumonia, the glucose in pleural effusion is >3.3 mmol/L, the PH is >7.2, and the pleural effusion can be absorbed after anti-infection treatment. Cord blood was collected from uninfected full-term newborns at Sun Yat-sen Memorial Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All participants in this study signed informed consent statements. This study was approved by the Ethics Review Committee of Sun Yat-sen University Cancer Center (No. GZR2018-182). The authenticity of this paper has been verified and key raw data have been uploaded to the Research Data Deposit (RDD) Public platform (www.researchdata.org.cn) with RDD number RDDB2401150003.
Antigens and antibodies
The following antibodies were used for cell surface and intracellular stainings: FITC-labeled anti-CD3 (#566783), APC-Cy7-labeled anti-CD4 (#557871), PE-labeled anti-CD69 (#555531), IL-2-PE (#569370), PerCP-labeled anti-CD69 (#340548), CD45RO-PerCP-Cy5.5 (#560607), PE-Cy7-labeled anti-CD45RA (#561216), APC-labeled anti-IFN-γ (#655933), TNF-α-PE-Cy7 (#557647), isotype control antibodies, and purified anti-CD3 (#555330) and anti-CD28 (#348040), which were purchased from Becton, Dickinson, and Co. (BD) Biosciences Pharmingen (San Jose, CA, USA).
Isolation and culture of mononuclear cells from pleural effusion and preparation of cord blood mononuclear cells (CBMCs)
Pleural effusion samples from BPE and MPE patients were extracted by pleural puncture. Pleural fluid mononuclear cells (PFMCs) were isolated by density gradient centrifugation of 400 ×g with Ficoll-Hypaque (Tianjin Haoyang Biological Manufacture, Tianjin, China) and centrifugation for 20 minutes. Cells were collected and washed twice with Hank’s balanced salt solution (HBSS) (Sigma-Aldrich, St. Louis, MO, USA) at 400 ×g for 8 minutes. CD3 T cells were negatively selected with anti-CD3 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) with a mean purity of 98% and measured by flow cytometry. Dead cells were stained with trypan blue to detect cell viability. Finally, the cells were suspended in Roswell Park Memorial Institute (RPMI) 1640 medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal calf serum (Sijiqing, Hangzhou, China), 100 U/mL penicillin, 100 g/mL streptomycin, 2 mM l-glutamine, and 50 M 2-mercaptoethanol (Life Technologies) at 1×106 cells/mL. Heparinized cord blood was thoroughly mixed with dextran 500 solution (GE Healthcare Bio-Sciences, Uppsala, Sweden) and cultured in a 5% CO2 incubator at 37 °C to remove the red blood cells. After 30 minutes, CBMCs were obtained by density gradient centrifugation with Ficoll-Hypaque.
Isolation of T-cell subsets and Th1 cell differentiation conditions
CD3+ T cells were negatively isolated from freshly isolated PFMCs and CBMCs using biotin-conjugated antibodies (Miltenyi Biotec). Cells were then sent through a magnetic column to collect T cells. Labeled cells were collected as CD3-deficient cells. The purity of T cells was determined as more than 98% by flow cytometry (FACS Calibur; BD Biosciences). CD4+ T cells were negatively isolated from PFMCs and CBMCs using a biotin-antibody mixture (Miltenyi Biotec). Unlabeled cells were collected as CD4+ T cells. The purity of CD4+ T cells was determined as more than 96% by flow cytometry. To induce Th1 cells, CD4+ T cells were purified from CBMCs and cultured with anti-CD3 and anti-CD28 + hIL-12 (5 ng/mL) and anti-hIL-4 (2 mg/mL). On day 5, cells were collected and washed with phorbol 12 myristate 13-acetate (PMA, 20 ng/mL; Sigma-Aldrich) + ionomycin (1 µg/mL; Sigma-Aldrich) for 1 and 3 days, respectively. In addition, 2×105 cells/well differentiated Th1 cells were re-stimulated with PMA + ionomycin. Culture supernatants were collected and analyzed by enzyme-linked immunosorbent assay (ELISA) for the production of IL-2 and TNF-α on day 1 and IFN-γ on day 3.
Cell transfection and luciferase assay
MiR-16-5p inhibitor (5'-CACCAAUAUUUACGUGCUGCUA-3'), mimic (5'-UAGCAGCACGUAAAUAUUGGCG-3'), and corresponding negative control (NC) (5'-UUCUCCGAACGUGUCACGUTT-3') were purchased from Guangzhou RiboBio Co. Ltd. (Guangzhou, China). Transfection was performed according to instructions with Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). Luciferase reporter gene assay verified the binding and interaction between miR-16-5p and the 5'-UTR of IFN-γ mRNA. IFN-γ mRNA fragments containing miR-16-5p binding sites were cloned into the pMIR-REPORT luciferase reporter vector. Luciferase reporter vectors and miR-16-5p mimic expression vectors or miR-16-5p inhibitors were co-transfected into naïve CD4+ T cells, and luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
miRNA microarray assay and miRNA verification by reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from PFMC and CBMC using TRIzol (Invitrogen) according to the manufacturer’s instructions. Total RNA was quantified using high sensitivity Qubit Assays with the Qubit 4.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). miRNA microarray analysis was performed by the service provider (LC Sciences, Houston, TX, USA) using the methods described previously (29,30). RT-PCR was used in DNA thermal cycler apparatus (Applied Biosystems 2720; Thermo Fisher Scientific) under the following conditions: 95 °C for 5 minutes, 94 °C for 30 seconds, 62 °C for 30 seconds, and 72 °C for 60 seconds performed 25–30 cycles, then 72 °C for 5 minutes. The following primer pairs (Takara, Kusatsu, Shiga, Japan) were used: miR-16-5p forward 5'-TAGCAGCACGTAAATATTGGCG-3' and reverse 5'-TGCGTGTCGTGGAGTC-3'; miR-30e forward 5'-GTGCCTCACTGCGTCTC-3', and reverse 5'-GAAAGCCGGTGCGTAGCTG-3'; miR-320e forward 5'-GGGAAAGCTGG GTTGAGAA-3' and reverse 5'-GTCGGTGTCGTGGAGTCGTT-3'; miR-22 forward 5'-GGGGGATCCCTGGGGCAGGACCCT-3', and reverse 5'-GGGGAATTCAACGTATCATCCACCC-3'; miR-let-7a forward 5'-CCTGG ATGTTCTCTTCACTG-3', and reverse 5'-GCCTGGATGCAGACTTTTCT-3'; miR-2116 forward 5'-AATCCTATGCCAAGAACTCCC-3', and reverse 5'-CTCTACAGCTATATTGCCAGCCA-3'; miR-208a forward 5’-GTCATCTAGAAAGCTTGATGCAGGAAAGAGCTTTGG-3' and reverse 5'-TGACAGATCTCAGCTGACATCCTCTAGGCT; miR-3074 forward 5'-ACCATTCCTGCTGA ACTGAG-3', and reverse 5'-GCGAGCACAGAATTAATACGAC-3'; U6 forward 5'-GACCGAGTGTAGCAAGG-3', reverse 5'-GTTCTTCCGAGAACATATAC-3’.
miRNA expression in each sample was normalized to internal control (U6) and was expressed by comparison threshold (Ct) cycling method (2−ΔΔCt). Differences between groups were assessed using the paired Student’s t-test or the Mann-Whitney test.
ELISA
Culture supernatants were collected and assayed for the production of IFN-γ, IL-2, and TNF-α (BD Biosciences Pharmingen) by ELISA according to the manufacturer’s instructions. The sensitivity of ELISA kit for IFN-γ, IL-2, and TNF-α was 7.8 pg/mL.
Cell surface/intracellular cytokine staining and flow cytometric analysis
The surface stain was washed twice with phosphate-buffered saline (PBS) buffer containing 0.1% bovine serum albumin (BSA) and 0.05% sodium azide. The cells were then incubated with anti-CD3, anti-CD4, anti-CD69, anti-CD45RO, and anti-CD45RA surface markers in 4 °C darkness for 30 minutes. The cells were washed twice and fixed with 1% paraformaldehyde before collection. Intracellular cytokines were detected with anti-CD3, anti-CD28, brefeldin A (10 mg/mL; Sigma-Aldrich) in the last 6 hours of the experiment. After stimulation, cells were washed twice with PBS buffer containing 0.1% BSA and 0.05% sodium azide. The cells were then incubated with anti-CD3, anti-CD4, and anti-CD69 surface markers in 4 °C darkness for 30 minutes. The cells were washed twice, fixed with 4% paraformaldehyde, then permeated, and underwent intracellular cytokine staining. Flow cytometry was performed by BD FACSCalibur and FACSAria II (BD Biosciences), and data analysis was performed by FlowJo software (TreeStar, San Carlos, CA, USA).
Western blotting
Cells were washed lysed in buffer containing 1% (v/v) Triton X-100, 1 mM ethylenediamine tetraacetic acid (EDTA), 50 mM Tris (pH 7.5), 10% (v/v) glycerol, 150 mM NaCl, and a protease inhibitor. Cell lysates were isolated on 10% sodium dodecyl sulfate (SDS) gels and transferred to polyvinylidene fluoride (Millipore, Billerica, MA, USA). After the membrane was blocked, rabbit polyclonal antibody (pAbs) T-bet [Cell Signaling Technology (CST), Danvers, MA, USA] and goat anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA) coupled with horseradish peroxidase were used for immunoassay.
A rat monoclonal antibody (Santa Cruz Biotechnology) was used as a loading control. Visualization was performed using the enhanced chemiluminescence (ECL) western blotting analysis system (CST) according to the manufacturer’s instructions.
Statistical analysis
Statistical differences were measured by the unpaired Student’s t-test, two-tailed (Prism 5.0 software; GraphPad, San Diego, CA, USA). The Kruskal-Wallis test and Dunn’s multiple comparison test were used to assess the differences between groups. P<0.05 was a significant difference, P<0.01 was a very significant difference, and P<0.001 was an extremely significant difference.
Results
Patient characteristics
A total of 30 NSCLC patients with MPE and 30 patients with BPE were firstly analysed. The clinical characteristics of all patients are shown in Table 1. The median ages of the patients with MPE and those with BPE were 57 (range, 39–80) and 51 (range, 29–83) years. There were 17 males (56.67%) and 13 females (43.33%) with MPE and 16 males (53.33%) and 14 females (46.67%) with BPE. There were 14 cases (46.67%) of lung adenocarcinoma and 16 cases (53.33%) of lung squamous cell carcinoma among patients with MPE, and 20 cases (66.67%) of tuberculosis and 10 cases (33.33%) of pneumonia among patients with BPE. MPE was diagnosed in cytology or pleural biopsy, and BPE was diagnosed in biochemistry or pleural biopsy (Table 1).
Table 1
| Variable | MPE | BPE |
|---|---|---|
| Patients, n | 30 | 30 |
| Age (years), median [range] | 57 [39–80] | 51 [29–83] |
| Gender, n (%) | ||
| Male | 17 (56.67) | 16 (53.33) |
| Female | 13 (43.33) | 14 (46.67) |
| MPE, n (%) | ||
| Adenocarcinoma | 14 (46.67) | – |
| Squamous cell carcinoma | 16 (53.33) | – |
| BPE, n (%) | ||
| Tuberculous | – | 20 (66.67) |
| Parapneumonic | – | 10 (33.33) |
| Diagnostic method, n (%) | ||
| Biochemistry | – | 24 (80.00) |
| Cytology | 25 (83.33) | – |
| Pleural biopsy | 5 (16.67) | 6 (20.00) |
NSCLC, non-small cell lung cancer; MPE, malignant pleural effusion; BPE, benign pleural effusion.
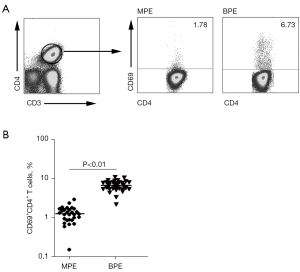
The expression level of CD4+CD69+ T cells in MPE was significantly lower than that in BPE
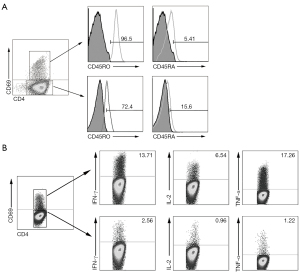
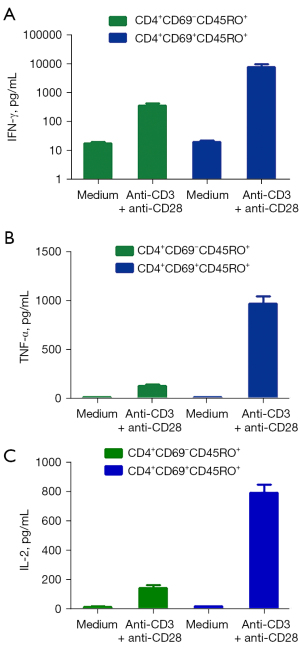
In order to investigate phenotypic markers of memory CD4+ T cells in PFMCs from MPE with advanced NSCLC, flow cytometry was used to detect the phenotype of memory CD4+ T cells in advanced NSCLC with MPE and lung disease with BPE. We found that the level of CD4+ T cell expression activation molecule CD69 in MPE was significantly lower than that in BPE (Figure 1A, 1B). Importantly, a total of 96.5% of CD4+CD69+ T cells expressed CD45RO (a major marker of memory T cells), and only 5.41% of CD4+CD69+ T cells expressed CD45RA. At the same time, we also found that 72.4% of CD4+CD69− T cells also expressed CD45RO, and 15.6% of CD4+CD69− T cells expressed CD45RA (Figure 2A). In order to further elucidate the immune role of memory CD4+ T cells in the progression of lung cancer, these cells were stimulated by anti-CD3 and anti-CD28, and the cytokines expression levels were detected by flow cytometry. We found that CD4+CD69+ T cells highly expressed Th1 cytokines such as IFN-γ, TNF-α, and IL-2, whereas CD4+CD69− T cells rarely expressed IFN-γ, TNF-α, and IL-2 after anti-CD3 and anti-CD28 stimulation (Figure 2B). Meanwhile, we used flow cytometry to sort two cell subsets, including CD4+CD69+CD45RO+ T cells and CD4+CD69−CD45RO+ T cells. After being stimulated by anti-CD3 and anti-CD28, the supernatant was collected and the production of cytokines including IFN-γ, TNF-α, and IL-2 was measured by ELISA. We found that CD4+CD69+CD45RO+ T cells but not CD4+CD69−CD45RO+ T cells significantly enhanced the production of IFN-γ, TNF-α, and IL-2 after anti-CD3 and anti-CD28 stimulation (Figure 3A-3C).
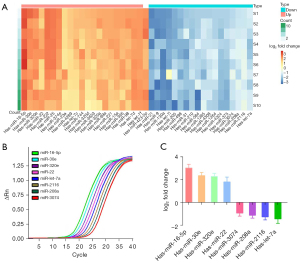
The expression level of miR-16-5p in MPE was significantly higher than that in BPE
In order to investigate miRNA that play a role in memory CD4+ T cell activation, a flow cytometry was used to sort CD4+CD69+ T cells in advanced NSCLC with MPE and benign lung disease with BPE. The expression of miRNAs in CD4+CD69+ T cells was detected by miRNA microarray. Our results showed that there were 22 miRNAs with significantly up-regulated expression in CD4+CD69+ T cells of NSCLC with MPE [fold change (FC) >2], among which miR-16-5p was the most significantly up-regulated. At the same time, there were 18 miRNAs with significantly down-regulated expression (FC <0.5) (Figure 4A). In order to verify the detection results of miRNA microarray, the expression of up-regulated miR-16-5p, miR-30e, miR-320e, and miR-22 and down-regulated miR-let-7a, miR-2116, miR-208a, and miR-3074 were detected by RT-PCR. We found that the results of the two methods were consistent (Figure 4B,4C).
MiR-16-5p directly targeted IFN-γ
In order to find the target genes of miRNA, three online databases were used, including Pic Tar (available at https://pictar.mdc-berlin.de), miRanda 3.0 (available at https://www.cs.kent.ac.uk/people/staff/dat/miranda), and TargetScan 3.1 (available at https://www.targetscan.org) to analyze and predict the target genes of the four miRNAs with the highest up-regulated expression. Our results showed that although miR-30e, miR-320e, and miR-22 all had certain regulatory effects on IFN-γ, miR-16-5p can bind to both the IFN-γ promoter and its 5'UTR. Therefore, we speculated that miR-16-5p might directly regulate the expression of IFN-γ.
To confirm the reliability of prediction of target genes, naïve CD4+ T cells were isolated from CBMCs. Cells were transfected with the IFN-γ wild-type (IFN-γWT-luciferase) or mutant (IFN-γMut-luciferase) luciferase reporter vector along with miR-16-5p mimic, miR-16-5p inhibitor, or negative control for 48 hours. Our results clearly suggested that after transfecting naïve CD4+ T cells with miR-16-5p mimics, the expression level of IFN-γ was significantly reduced, and there was no significant change in miR-16-5p inhibitors or mutants (IFN-γMut-luciferase) (Figure 5A). It was further indicated that there was a binding relationship between miR-16-5p and IFN-γ. Western blot analysis showed that overexpression of miR-16-5p could reduce IFN-γ protein.
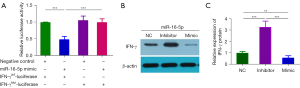
Similarly, inhibition of miR-16-5p expression could increase the level of IFN-γ protein (Figure 5B,5C). Our results showed that IFN-γ gene was the target gene directly acted by miR-16-5p, and miR-16-5p could inhibit the expression of IFN-γ gene.
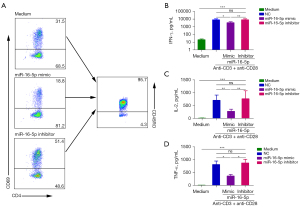
miR-16-5p inhibited Th1 cell differentiation and cytokine production
In order to test the effect of miR-16-5p on the functional response of CD4+ T cells from CBMCs, miR-16-5p mimic and miR-16-5p inhibitor were transfected into naïve CD4+ T cells. The cells were then cultured under Th1-polarized conditions of anti-CD3+anti-CD28, anti-hIL-4 and hIL-12. After 5 days, cells were collected, washed, and stimulated with PMA + ionomycin. Th1 cell development was notably blunted when miR-16-5p mimic was added to Th1 cell differentiation culture conditions. Conversely, in the presence of miR-16-5p inhibitor, the number of Th1 cells increased significantly (Figure 6A). Consistent with fluorescence-activated cell sorting (FACS) analysis, the results of ELISA showed that naïve CD4+ T cells cultured under Th1-polarized conditions with miR-16-5p mimic significantly reduced the production of IFN-γ, IL-2, and TNF-α (P<0.001). The addition of miR-16-5p inhibitors under Th1 polarization conditions significantly increased the production of IFN-γ, IL-2, and TNF-α (Figure 6B-6D). These results established the view that miR-16-5p had a direct inhibitory effect on Th1 cell differentiation and cytokines production.
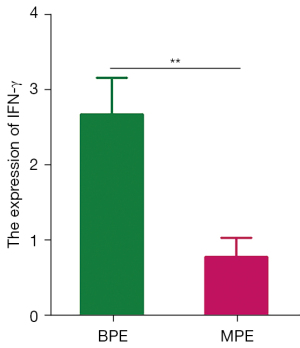
IFN-γ was down-regulated in CD4+CD69+ T cells in MPE and IFN-γ could promote the expression of T-bet protein in CBMCs
IFN-γ expression was assessed by RT-PCR in BPE or NSCLC with MPE. The results showed that IFN-γ was significantly decreased in MPE compared with that in BPE (Figure 7).
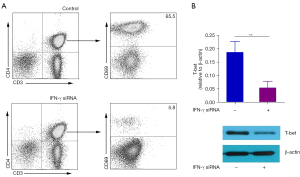
To investigate the role of IFN-γ in the differentiation of CD4+ T cells, we treated the naïve CD4+ T cells with or without small interfering RNA (siRNA) of IFN-γ.
Our results showed that naïve CD4+ T cells down-regulated by siRNA of IFN-γ differentiated into fewer memory CD4+CD69+T cells than naïve CD4+ T cells without siRNA of IFN-γ (Figure 8A). At the same time, the results of western blot showed that naïve CD4+ T cells down-regulated by IFN-γ siRNA expressed lower levels of T-bet than naïve CD4+ T cells (Figure 8B). It is suggested that miR-16-5 may be dependent on IFN-γ to regulate the differentiation of memory CD4+CD69+ T cells in MPE of NSCLC.
Discussion
The prognosis of NSCLC remains poor although the therapeutic option is dramatically changing (31). The immune system plays an important role in the development of NSCLC, and CD4+ T cells are critical for anti-tumor immune responses (32,33). The complex interactions among different immune cells have important functions in the development of MPE. Immune cells in MPE exhibit many transcriptional features that are enriched in regulatory T cells, B cells, macrophages, and dendritic cells. Cytotoxic T, helper T, regulatory T and Th cells express multiple immune checkpoints or co-stimulatory molecules. Cell-cell interaction analysis found that regulatory B cells interacted more with CD4+ T cells than CD8+ T cells. Macrophages showed transcriptional heterogeneity, which was consistent with M2 polarization. In addition, immune cells in MPE exhibit a general up-regulation of glycolytic pathways associated with the hypoxic microenvironment (34-36). However, the mechanism behind CD4+ T cell differentiation in NSCLC with MPE remains poorly understood. In order to identify and characterize several functional CD4+ T-cell subsets in malignant pleural effusion of NSCLC patients, flow cytometry was used to detect the phenotype of memory CD4+ T cells in advanced NSCLC with MPE. Our results showed that a total of 96.5% of CD4+CD69+ T cells expressed CD45RO, and only 5.41% of CD4+CD69+ T cells expressed CD45RA. We also found that 72.4% of CD4+CD69− T cells expressed CD45RO, and 15.6% of CD4+CD69− T cells expressed CD45RA. After stimulated by anti-CD3 and anti-CD28, CD4+CD69+ T cells, but not CD4+CD69−T cells highly express Th1 cytokines such as IFN-γ, TNF-α, and IL-2. Meanwhile, we used flow cytometry to sort CD4+CD69+CD45RO+ T cells and CD4+CD69−CD45RO+ T cells. The supernatant was collected and the production of cytokines including IFN-γ, TNF-α, and IL-2 was measured by ELISA after anti-CD3 and anti-CD28 stimulation. We found that CD4+CD69+CD45RO+ T cells but not CD4+CD69−CD45RO+ T cells significantly enhanced the production of IFN-γ, TNF-α, and IL-2 after anti-CD3 and anti-CD28 stimulation.
MiR-16-5p is a microRNA that plays an important role in the development of various malignancies, including neuroblastoma, osteosarcoma, hepatocellular carcinoma, cervical cancer, breast cancer, brain tumor, gastrointestinal cancer, lung cancer and bladder cancer (37). For example, in neuroblastoma, inhibition of miR-16-5p up-regulation of circ-CUX1 is associated with advanced tumor, node, metastasis (TNM), low differentiation grade and lymph node metastasis (38). MiR-16-5p has been shown to be underexpressed in patients with breast cancer. Moreover, patients with low expression of miR-16-5p had lower survival compared to patients with high expression of miR-16-5p (39). In addition, expression levels of AGAP2-AS1 and miR-16-5p are associated with clinical parameters and poor prognosis in patients with hepatocellular carcinoma (40).
In this study, we identified miR-16-5p as a critical regulatory factor in NSCLC. Our study revealed that the expression of miR-16-5p was significantly increased in CD4+CD69+ T cells in NSCLC with MPE, and downregulated the expression of IFN-γ, which is essential for Th1 polarization and memory Th cell differentiation.
Previous research has shown that miR-16-5p is involved in the regulation of several crucial pathways, including the control of cell proliferation, differentiation, and apoptosis. Overexpression of miR-16-5p can inhibit proliferation, migration, and invasion of osteosarcoma cells, and enhance the therapeutic effect of cisplatin (41). A study has shown that overexpression of miR-16-5p can inhibit the proliferation, invasion, and migration of chordoma cells in vitro and in vivo, and is associated with up-regulation of E-cadherin expression and down-regulation of N-cadherin and vimentin expression (42). Another study showed that miR-16-5p, which is lowly expressed in breast cancer tissues, can inhibit breast cancer by inhibiting the NF-κB pathway and decreasing AKT3 (39). Thus, it is clear that miR-16-5p has several biological functions depending on the cellular context.
Our study provides evidence that miR-16-5p is a crucial factor in NSCLC with MPE, as it specifically inhibits IFN-γ regulated memory Th cell differentiation. Our results indicate for the first time that miR-16-5p may serve as a potential therapeutic target for the treatment of NSCLC with MPE. The data presented in this study imply that miRNAs, such as miR-16-5p, play a significant role in the regulation of the immune response against NSCLC. Our results suggest that patients with advanced NSCLC with MPE who express the memory CD4+CD69+CD45RO+ T cell phenotype are most likely to benefit from our findings. At the same time, we will continue to expand the number of samples in the following studies to further verify the mechanism of miR-16-5p’s involvement in regulating the formation and differentiation of memory CD4+ T cells in lung cancer MPE, analyze its possible signal transduction pathway, and reveal the specific mechanism of immune escape of tumor cells in lung cancer MPE. This study provides scientific basis for guiding the formation and regulation mechanism of immune memory in the pathogenesis of MPE in lung cancer and seeking new therapeutic targets.
There are four limitations in the current study. First, PFMCs isolated by density gradient centrifugation may not represent the bulk tumor immune microenvironment because the supernatant of MPE is ignored in the study. The tumor immune microenvironment of MPE may have difference between in-vitro and in-vivo. Second, the role of miR-16-5p is shown by only in-vitro experiment. The role of miR-16-5p should be verified by another model animal. Third, the mechanism of miR-16-5p to regulate IFN-γ is not clarified. The actual affinity of miR-16-5p to IFN-γ is not shown although the prediction of it is enough high. Fourth, the sample size of this study is limited, and further research with a large sample size will be carried out under appropriate conditions in the later stage.
Conclusions
Our study provides important insights on the role of miR-16-5p in NSCLC. The findings could be useful for the development of novel therapeutic strategies.
Acknowledgments
We thank Dr. Kuanxia Wei for her critical revision of the manuscript for important intellectual content.
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-505/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-505/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-505/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-505/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Review Committee of Sun Yat-sen University Cancer Center (No. GZR2018-182) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ruan X, Sun Y, Wang W, et al. Multiplexed molecular profiling of lung cancer with malignant pleural effusion using next generation sequencing in Chinese patients. Oncol Lett 2020;19:3495-505. [Crossref] [PubMed]
- Oja AE, Piet B, van der Zwan D, et al. Functional Heterogeneity of CD4(+) Tumor-Infiltrating Lymphocytes With a Resident Memory Phenotype in NSCLC. Front Immunol 2018;9:2654. [Crossref] [PubMed]
- DeLong P, Carroll RG, Henry AC, et al. Regulatory T cells and cytokines in malignant pleural effusions secondary to mesothelioma and carcinoma. Cancer Biol Ther 2005;4:342-6. [Crossref] [PubMed]
- Ryu JS, Ryu HJ, Lee SN, et al. Prognostic impact of minimal pleural effusion in non-small-cell lung cancer. J Clin Oncol 2014;32:960-7. [Crossref] [PubMed]
- Shinohara S, Takahashi Y, Komuro H, et al. New evaluation of the tumor immune microenvironment of non-small cell lung cancer and its association with prognosis. J Immunother Cancer 2022;10:e003765. [Crossref] [PubMed]
- Yi FS, Zhai K, Shi HZ. Helper T cells in malignant pleural effusion. Cancer Lett 2021;500:21-8. [Crossref] [PubMed]
- Wheeler BD, Gagnon JD, Zhu WS, et al. The lncRNA Malat1 inhibits miR-15/16 to enhance cytotoxic T cell activation and memory cell formation. Elife 2023;12:RP87900. [Crossref] [PubMed]
- Ding M, Zhang C, Wang W, et al. Silica-exposed macrophages-secreted exosomal miR125a-5p induces Th1/Th2 and Treg/Th17 cell imbalance and promotes fibroblast transdifferentiation. Ecotoxicol Environ Saf 2023;267:115647. [Crossref] [PubMed]
- Bolandi Z, Hashemi SM, Abasi M, et al. In vitro naive CD4(+) T cell differentiation upon treatment with miR-29b-loaded exosomes from mesenchymal stem cells. Mol Biol Rep 2023;50:9037-46. [Crossref] [PubMed]
- Dong J, Huth WJ, Marcel N, et al. miR-15/16 clusters restrict effector Treg cell differentiation and function. J Exp Med 2023;220:e20230321. [Crossref] [PubMed]
- Peng M, Wang J, Tian Z, et al. Autophagy-mediated Mir6981 degradation exhibits CDKN1B promotion of PHLPP1 protein translation. Autophagy 2019;15:1523-38. [Crossref] [PubMed]
- Yan L, Liang M, Hou X, et al. The role of microRNA-16 in the pathogenesis of autoimmune diseases: A comprehensive review. Biomed Pharmacother 2019;112:108583. [Crossref] [PubMed]
- Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays 2014;36:617-26. [Crossref] [PubMed]
- Tian Z, Hua X, Zhu J, et al. ATG7 upregulation contributes to malignant transformation of human bronchial epithelial cells by B[a]PDE via DNMT3B protein degradation and miR-494 promoter methylation. Ecotoxicol Environ Saf 2023;263:115273. [Crossref] [PubMed]
- Lv M, Xu Y, Tang R, et al. miR141-CXCL1-CXCR2 signaling-induced Treg recruitment regulates metastases and survival of non-small cell lung cancer. Mol Cancer Ther 2014;13:3152-62. [Crossref] [PubMed]
- Wang W, Wang S, Pan L. Identification of key differentially expressed mRNAs and microRNAs in non-small cell lung cancer using bioinformatics analysis. Exp Ther Med 2020;20:3720-32. [Crossref] [PubMed]
- Zhang J, Nie R, Liu M, et al. A Novel Strategy for Identifying NSCLC MicroRNA Biomarkers and Their Mechanism Analysis Based on a Brand-New CeRNA-Hub-FFL Network. Int J Mol Sci 2022;23:11303. [Crossref] [PubMed]
- Wang T, Lv M, Shen S, et al. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS One 2012;7:e43268. [Crossref] [PubMed]
- Hu Y, Paris S, Barsoumian H, et al. A radioenhancing nanoparticle mediated immunoradiation improves survival and generates long-term antitumor immune memory in an anti-PD1-resistant murine lung cancer model. J Nanobiotechnology 2021;19:416. [Crossref] [PubMed]
- Goc J, Germain C, Vo-Bourgais TK, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res 2014;74:705-15. [Crossref] [PubMed]
- Umeshappa CS, Nanjundappa RH, Xie Y, et al. Differential requirements of CD4(+) T-cell signals for effector cytotoxic T-lymphocyte (CTL) priming and functional memory CTL development at higher CD8(+) T-cell precursor frequency. Immunology 2013;138:298-306. [Crossref] [PubMed]
- Nakayama T, Hirahara K, Kimura MY, et al. CD4+ T cells in inflammatory diseases: pathogenic T-helper cells and the CD69-Myl9 system. Int Immunol 2021;33:699-704. [Crossref] [PubMed]
- Hayashizaki K, Kimura MY, Tokoyoda K, et al. Myosin light chains 9 and 12 are functional ligands for CD69 that regulate airway inflammation. Sci Immunol 2016;1:eaaf9154. [Crossref] [PubMed]
- Kohlmeier JE, Miller SC, Smith J, et al. The chemokine receptor CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity 2008;29:101-13. [Crossref] [PubMed]
- Sheng SY, Gu Y, Lu CG, et al. The distribution and function of human memory T cell subsets in lung cancer. Immunol Res 2017;65:639-50. [Crossref] [PubMed]
- Neurath MF, Finotto S. The emerging role of T cell cytokines in non-small cell lung cancer. Cytokine Growth Factor Rev 2012;23:315-22. [Crossref] [PubMed]
- Wu Y, Yuan M, Wang C, et al. T lymphocyte cell: A pivotal player in lung cancer. Front Immunol 2023;14:1102778. [Crossref] [PubMed]
- Malik PS, Raina V, Singh A, et al. Analysis of regulatory T cells in patients of NSCLC with malignant pleural effusion. J Clin Oncol 2012;30:e17511. [Crossref]
- Lin F, Li R, Pan ZX, et al. miR-26b promotes granulosa cell apoptosis by targeting ATM during follicular atresia in porcine ovary. PLoS One 2012;7:e38640. [Crossref] [PubMed]
- Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012;61:1447-53. [Crossref] [PubMed]
- Tan WL, Jain A, Takano A, et al. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol 2016;17:e347-62. [Crossref] [PubMed]
- Wang Y, Liu M, Zhang L, et al. Cancer CD39 drives metabolic adaption and mal-differentiation of CD4(+) T cells in patients with non-small-cell lung cancer. Cell Death Dis 2023;14:804. [Crossref] [PubMed]
- Lau D, Khare S, Stein MM, et al. Integration of tumor extrinsic and intrinsic features associates with immunotherapy response in non-small cell lung cancer. Nat Commun 2022;13:4053. [Crossref] [PubMed]
- Huang ZY, Shao MM, Zhang JC, et al. Single-cell analysis of diverse immune phenotypes in malignant pleural effusion. Nat Commun 2021;12:6690. [Crossref] [PubMed]
- Lin H, Tong ZH, Xu QQ, et al. Interplay of Th1 and Th17 cells in murine models of malignant pleural effusion. Am J Respir Crit Care Med 2014;189:697-706. [Crossref] [PubMed]
- Wu XZ, Shi XY, Zhai K, et al. Activated naïve B cells promote development of malignant pleural effusion by differential regulation of T(H)1 and T(H)17 response. Am J Physiol Lung Cell Mol Physiol 2018;315:L443-55. [Crossref] [PubMed]
- Ghafouri-Fard S, Khoshbakht T, Hussen BM, et al. A review on the role of mir-16-5p in the carcinogenesis. Cancer Cell Int 2022;22:342. [Crossref] [PubMed]
- Zhang X, Zhang J, Liu Q, et al. Circ-CUX1 Accelerates the Progression of Neuroblastoma via miR-16-5p/DMRT2 Axis. Neurochem Res 2020;45:2840-55. [Crossref] [PubMed]
- Ruan L, Qian X. MiR-16-5p inhibits breast cancer by reducing AKT3 to restrain NF-κB pathway. Biosci Rep 2019;39:BSR20191611. [Crossref] [PubMed]
- Liu Z, Wang Y, Wang L, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38:194. [Crossref] [PubMed]
- Gu Z, Li Z, Xu R, et al. miR-16-5p Suppresses Progression and Invasion of Osteosarcoma via Targeting at Smad3. Front Pharmacol 2020;11:1324. [Crossref] [PubMed]
- Zhang H, Yang K, Ren T, et al. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis 2018;9:680. [Crossref] [PubMed]
(English Language Editor: J. Jones)


