
Oncogene-addicted solid tumors and microbiome—lung cancer as a main character: a narrative review
Introduction
Background
Over the past decade, there has been a notable focus on the microbiome, driven by increasing evidence linking microbiota to the natural history of tumors and its relationship with lifestyle, carcinogenesis, and responses to cancer treatments such as immune checkpoint blockers (ICB), and the severity of immune-related adverse effect (1-3). Bacteria, viruses and fungi colonize the lower respiratory tract of patients and healthy individuals. These microorganisms are thought to play a specific role, including resistance to pathogens, activation of the immune system and uptake of nutrients (4). Dickson et al. described that Prevotella, Veillonella and Streptococcus are the main genera in lung tissue of healthy individuals, although they appeared in low quantity. Compared to the gut, lung microbiota is known to vary more over time, with a consistent spatial variation (5,6). Three factors have been described to impact the composition of lung microbiota: (I) introduction of microbiomes into the airway; (II) elimination of microbiomes from the airway; and (III) regional conditions that influence microbial growth. These conditions, influenced by environmental factors such as air pollution or cigarette smoking, are the most determinant during disease. Of note, particulate matter (PM) present in the air, derived from fossil fuel combustion and pollution, has been recently associated to increased risk of EGFR and KRAS-mutant non-small lung cancer cell (NSCLC). As a result, the association between oncogenic drivers’ pathways has become more intriguing, and the possibility of an impact of environmental pollution on lung microbiota is increasingly plausible. PM-associated bacteria, mainly represented by Actinobacteria and Proteobacteria, usually appear enriched in patients with chronic pulmonary obstructive disease (COPD) exacerbations and asthma, further strengthening the link between PM and disease (7-9). Other determinants, such as seasonal variations and ambient temperature, may also alter the microbiota. For example, a study of PM in China described a predominance of Proteobacteria, Bacteroidetes, Actinobacteria, Cyanobacteria, and Firmicutes, attributing these findings to the presence of PM2.5, air humidity, and tropical climate of the region (10). As regards cigarette smoking, responsible for nearly 80% of this highly preventable disease (11,12), it has been described to have a strong effect on microbial composition as it contains potential respiratory pathogens, such as Actinobacter, Clostridium, Klebsiella, Pseudomonas and Serratia. Moreover, it appears to enhance bacterial adhesion and alter host immune response, thus leading to airway dysbiosis due to chronic inflammation (13). Overall, these findings highlight a robust connection between the lung microbiome, environmental factors, and the inflammatory changes that increase vulnerability to diseases. The growing prominence of oncogene-addicted tumors in non-smokers might be attributed to external disruptors beyond cigarette smoking. Host and environmental factors, including spatial, temporal, and compositional variations, influence the lung microbiota, thus creating a dynamic microbial system within the lungs.
Rationale and knowledge gap
It must be highlighted that although “microbiome” and “microbiota” are sometimes used interchangeably, key differences distinguish both terms. The microbiome is defined as the complete set of genomes from all microorganisms in a given environment, comprising not only the community of microorganisms but also their structural elements, metabolites, and the prevailing environmental conditions. In contrast, the microbiota refers specifically to the living microorganisms present in a defined environment, such as the microbiota found in the oral and gut regions (14). This distinction is crucial for understanding the link with the host and tumor cells, as well as for the development of potential therapeutic strategies targeting the microbiota. For example, preclinical studies in mice have shown that changes in microbiota composition after antibiotic administration confer resistance to ICB, a phenomenon that can be reversed after fecal microbiota transplant (FMT) or co-housing, or with the use of probiotics like Bifidobacterium (15,16). In humans, several large series have shown unfavorable outcomes of patients under ICB while receiving antibiotics (17-19). However, while interaction between microbiota and ICBs, as well as chemotherapeutic agents, has already been well elucidated in lung cancer and other solid tumors such as melanoma, evidence regarding tyrosine kinase inhibitors (TKIs) is lacking.
Objective
The present review aims to shed light on the link between the composition of microbiota, the immune system, oncogenic drivers and targeted therapies in lung cancer scenario. Our objective is to understand the complex interplay between microbiota composition and oncogene-addiction, in order to generate hypothesis that might enhance therapeutic outcomes. We present this article in accordance with the Narrative Review reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-216/rc).
Methods
This narrative review consists of previously published articles, searched in the PubMed and Google Scholar databases. The terms used to identify relevant data included “oncogene addiction”, “lung cancer”, “MAPK”, “EGFR”, “KRAS”, “microbiome”, “airway microbiota”, “gut microbiota”, “tyrosine kinase inhibitors”. Articles written in English published from 2013 to 2024 were reviewed in detail, and additional, relevant articles were included to deepen information retrieved. The search strategy is summarized in Table 1.
Table 1
| Items | Specification |
|---|---|
| Date of search | November 2022 to May 2024 |
| Database and other source searched | PubMed, Google Scholar |
| Search terms used | “oncogene addiction”, “lung cancer”, “MAPK”, “EGFR”, “KRAS”, “microbiome”, “airway microbiota”, “gut microbiota”, “tyrosine kinase inhibitors” |
| Timeframe | From 2013 to May 2024 |
| Inclusion criteria | Full-text English published articles were included |
| Selection process | M.G. and M.L.M. conducted the initial selection independently, and obtained consensus with the authors to include relevant information |
Microbiota, carcinogenesis, tumor microenvironment and immune system
The Human microbiota, described by Wang et al. as an “essential organ”, comprises nearly 100 trillion symbiotic microorganisms (20). There is evidence suggesting that microbiota generates a state of chronic inflammation, thus predisposing to various types of cancer. The impact of microbiota on patients’ responses to chemotherapy, radiotherapy, immune checkpoint blockade (ICB), and targeted therapy depends on its composition and its interactions with the host, the immune system, and the TME (21-24).
Microbiota and carcinogenesis
The role of the microbiota in carcinogenesis, response to treatments and appearance of adverse effects has been extensively studied in the last 20 years (25,26). Knippel et al. described that bacteria secrete toxins conducting direct DNA damage, establish an inflamed environment by the generation of metabolites, and maintain a chronic infectious state that potentiates immunosuppressive responses. This author also highlighted three hypothesis that may explain bacterial involvement in carcinogenesis: (I) the driver-passenger model, where commensal bacteria coordinate with a single group of tumorigenic bacteria to promote tumorigenesis; (II) the keystone theory that states that a single bacterium can favor the colonization of additional pro-carcinogenic bacteria thus leading to carcinogenesis; and (III) the hit-and-run model that supports the concept of a temporary colonization by carcinogenic bacteria that favors tumorigenesis (27,28). This inflammatory, pro-carcinogenic environment not only affects tumor initiation, promotion, invasion and metastasis, but also alters immune surveillance and responses to therapy. Pathways that converge to this tumorigenic state include secretion of immunosuppressive cytokines and chemokines, activation of oncogenic pathways such as RAS or MYC, and stimulation of mechanisms that favor proliferation and invasion, including angiogenesis and epithelial-mesenchymal transition (EMT) (29). Overall, microbial effect on tumorigenesis is a well-known mechanism established in various types of cancer. Examples of this include the link between Helicobacter pylori in gastric cancer, human papillomavirus (HPV) in cervical cancer, and Streptococcus bovis in colorectal cancer (CRC) (30,31).
Microbiota and the immune system
The relationship between the immune system and the microbiota has been on the spotlight in the last few years, mainly due to the interesting findings that link microbiota composition to efficacy and toxicity of ICB (32,33). This appears to be related to the fact that the microbiota helps to maintain immune homeostasis. Wu et al. reviewed the role of the microbiota on innate and adaptive immunity, highlighting its contribution on the development of antigen presenting cells (APC) and neutrophils, on the immunomodulatory role of intestinal epithelial cells, on the maturation and maintenance of CD4+ and CD8+ T cells, as well as on the production of cytokines and immunoglobulin A (34). Microbiota modulates immunity inducing either an immunosuppressive or an anti-tumor environment by altering neutrophil migration and function, T cell differentiation, and cytokine secretion (35). An example of this is the association between neutrophil-to-lymphocyte ratio (NLR) and microbiota. This index has been correlated with prognosis in cancer, as well as cardiovascular or inflammatory diseases (36,37). Studies have shown lower NLR has been also associated to a greater diversity of gut microbiota; certain species such as Bacteroides eggerthii have been linked to higher NLR and thus worse prognosis (38-40). A preclinical study by Sivan et al. found that mice had different anti-tumor responses depending on microbiota composition. In this study, Bifidobacterium was associated to augmented dendritic cell function and activation of CD8+ T cell response in TME (15).
As a result, the microbiota appears to play a critical role in shaping and regulating immune responses, as well as impacting on the development of immune-mediated disorders. These interactions can help regulate composition of commensal and homeostatic microbiota, or even promote a tolerigenic immune environment that leads to carcinogenesis and various diseases (41).
Microbiota and tumor microenvironment
TME plays a key role in tumor maintenance and progression (42). Dysbiosis, defined as the “change to the composition of resident commensal communities relative to the community found in healthy individuals”, has been associated with the appearance of multiple diseases and the induction of an immunosuppressive phenotype by stimulation of M2 macrophages, which suppress cytotoxic T cell response and promote tumor growth and metastasis (43,44). This inflammatory microenvironment, together with the secretion of microbial metabolites, stimulate angiogenesis and promotes tumor growth and progression (45). Gut microbiota has been also described to participate in immune evasion by secretion of cytokines, promotion of immunosuppressive function of myeloid derived suppressor cells (MDSC) and regulatory-T (Treg) cells, recruitment and differentiation of tumor associated macrophages (TAMs) and neutrophils, and down-regulation of CD8+ T-cell infiltration (46). Furthermore, certain intratumoral microbes such as Bifidobacterium, have been related to CD47 targeted immunotherapy response, showing that microbiota composition might be fundamental not only for cancer progression but also for therapy response (47).
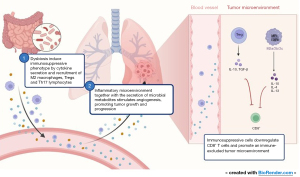
Overall, evidence has clearly sought to understand the role of microbiota in health and disease. Microbiota interacts with the immune system by modulating immune responses, and the host’s immune system helps maintain microbiome homeostasis. Dysbiosis can lead to a dysregulation of this delicate balance, thus promoting inflammation, immunosuppression and carcinogenesis. This intricate network of immune cells, TME components and their relationship with the human microbiome still needs to be further explored, mainly because it can deepen our understanding on carcinogenesis and help us in developing better therapeutic strategies. Figure 1 summarizes the impact of microbiome in TME and immune reprogramming.
Microbiota in lung cancer patients
Lung cancer is the leading cause of cancer death worldwide, according to the World Health Organization (WHO) (48). As a result, deepening our understanding of the microbiome in this subset of patients might help us comprehend disease progression and treatment response, in order to discover potential therapeutic interventions. Figure 2 illustrates the available evidence of microbiota composition in lung cancer patients when compared to healthy subjects.
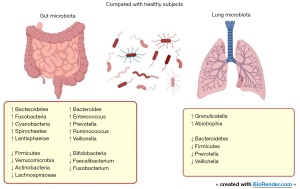
Concerning lung microbiota, Gomes et al. described an abundance of Proteobacteria in lung cancer patients, and correlated the presence of certain Enterobacteriaceae to worse survival. A distinct pattern was found in this study according to histology, whether adenocarcinoma, squamous cell or SCLC. An abundance of Acinetobacter, Propionibacterium, Phenylobacterium, Brevundimonas and Staphylococcus was found in adenocarcinoma subtype, while squamous cell carcinoma had a predominance of Enterobacter, Serratia, Kluyvera, Morganella, Achromobacter, Capnocytophaga and Klebsiella (49). Ramírez-Labrada et al. also reported several differences between healthy samples and tumor tissue, with an enrichment of Granulicatella, Abiotrophia, and Streptococcus in tumor tissue, and a predominance of Bacteroidetes, Firmicutes, Prevotella, Veillonella, and Streptococcus in healthy lungs (50). In addition, Greathouse et al. described an increase in diversity and richness in lung cancer samples as compared with normal tissue. An abundance of Acidovorax and Klebsiella, from phylum Proteobacteria, was found in smokers as well as in patients diagnosed with squamous cell carcinoma, as compared with non-smokers or adenocarcinoma subtype (51). Zhou et al. described an abundance of Bacteroidetes, Fusobacteria, Cyanobacteria, Spirochaetes, and Lentisphaerae, and lower levels of Firmicutes and Verrucomicrobia when compared to the control group (52). Similarly, other studies described an abundance of Firmicutes phylum in healthy lung tissue compared to controls, while associating certain families, including Lachnospiraceae and Ruminococcaceae, with reduced disease-free survival (53,54). This suggests that not only intratumoral bacteria, but also microbiota in healthy tissue, might play a role in recurrence.
Concerning intratumoral microbiota, Nejman et al. carried out a comprehensive analysis of 1,010 tumor samples of seven cancer types, including lung cancer, and 516 adjacent tissue normal samples. Bacteria were predominantly localized intracellularly, in cancer cells and immune cells, rising the hypothesis of whether they might play a role in carcinogenesis and tumor immunity. They also described an enrichment of Proteobacteria in lung cancer samples of smoker patients. This phylum was found to be associated to pathways that degrade chemicals from cigarette smoke, suggesting that smokers may present a specific environment that induces the presence of certain bacteria (55).
Overall, the available evidence highlights the differences in microbiota composition between lung cancer samples and healthy tissue, among different histologies, and between smokers and non-smokers. It is worth highlighting that these differences raise the question on whether this variability is the cause of carcinogenesis, or if it is the result of multifactorial conditions that involve treatments used, genetic background or even lifestyle and dietary habits. This reinforces the importance of the host, the environment, and external factors in this complex interplay.
Oncogene addicted tumors and microbiota
Oncogene addiction is defined as the phenomenon of cancer cell dependence on individual oncogenes to sustain the malignant phenotype, and one of the most representative players in this setting is lung cancer (56). Oncogenic driver genes are a key element of precision medicine. Considering nearly 41% of patients with lung cancer present with stage IV at first diagnosis, it is not noteworthy that targeted therapies have improved overall survival (OS) and tolerance to oncological treatments as compared with chemotherapy (57,58). Although there is data that link targetable oncogenic drivers and microbiome, evidence is scarce. Table 2 summarizes the current evidence.
Table 2
| Microbiome | Reference | Bacteria | Results |
|---|---|---|---|
| KRAS | Jin et al. (59) | Herbaspirillum and Sphingomonadaceae | Increased bacterial burden, decreased diversity. Increase in tumor burden. Treatment with antibiotics reduced tumor growth and high-grade lesions |
| EGFR and airway microbiome |
Zheng et al. (60) | Rhizopus oryzae, Natronolimnobius innermongolicus, Staphylococcus sciuri, Orenia marismortui, Burkholderia multivorans and Sinorhizobium | Differences found in lung microbiome according to age, gender, smoking and EGFR status |
| Huang et al. (61) | Bacteroidetes, Tenericutes, Sharpea, Prevotella, Porphyromonas, Parvimonas, Desulfovibrio, Mycoplasma, Actinobacillus, Dialister, and Eikenella | Alpha diversity between EGFR mutated and wild-type was similar. Differences in Beta diversity and in activated metabolic pathways | |
| EGFR and Gut microbiome | Otoshi et al. (62) | Blautia | Decreased levels of Bifidobacterium and Faecalibacterium compared to controls |
| Saifon et al. (63) | Bacteroidetes and Firmicutes | Higher alpha diversity in mutated. Similar Beta diversity between cohorts. Actinobacteria enrichment in patients with progressive disease after EGFR-TKI treatment |
|
| EGFR and intratumoral microbiome | Zhang et al. (64) | Serratia marcescens | Negative correlation to Haemophilus parainfluenzae. Serratia marcescens associated to better overall survival |
| MAPK pathway in other tumors |
Boonanantanasarn et al. (65) |
Enterococcus faecalis | Induction of EGFR pathway in patients with oral cancer. Production of H2O2 or EGF-like signals which stimulate cell proliferation |
| Wong et al. (66) | Helicobacter pylori | Increase in EGF protein and EGFR mRNA expression in the antral mucosa to promote injury repair and ulcer healing, but increasing risk of malignancy |
EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; MAPK, mitogen-activated protein kinases.
KRAS mutation-associated microbiota
A preclinical study analyzing the impact of microbiota in KRAS and p53-driven lung cancer adenocarcinoma showed germ-free mice exhibited delayed tumor growth and a smaller proportion of high-grade lesions (67). However, when these mice were exposed to 14 bacterial strains cultured from late-stage lung tumors, they experienced an increase in tumor burden, thus supporting the fact that certain bacteria play a role in tumor progression. Conversely, mice with specific pathogen-free conditions that displayed at first rapid tumor growth, were administered an antibiotic cocktail of ampicillin, neomycin, metronidazole, and vancomycin, which significantly reduced tumor growth and the occurrence of high-grade lesions. This response was associated with γδ T-cells activity, which secreted IL-17A, contributing to inflammation and anti-tumor immunity. In addition, they observed higher levels of IL-22, known for its tumor-promoting effect in colorectal adenocarcinoma, and a higher expression of IFN-γ, T-bet, and CD27, which play a role in tumor differentiation and maturation. Interestingly, tumor-bearing mice had an increase in bacterial burden but reduced diversity in the airway. This reduction of diversity was accompanied by an abundance of Herbaspirillum and Sphingomonadaceae in tumor samples, in contrast to the predominance of Aggregatibacter and Lactobacillus in healthy lungs (59,67). Sui et al. investigated the relationship of KRAS pathway with the intestinal microbiota in the tumor tissues of patients with CRC. The study found that the presence of KRAS was associated with a significant reduction in bacterial diversity and richness in the intestinal microbiota. Additionally, specific bacterial taxa such as Roseburia, Parabacteroides, Metascardovia, Staphylococcus and Bacillales were found to be more abundant in patients with KRAS mutations compared to those without (68). The expression of KRAS and BRAF have also been described to increase in the presence of Bacteroides fragilis and Fusobacterium nucleatum. Evidence highlights that these bacteria promote the development of CRC by modulating T-cell adaptive immunity and secreting cytokines (69).
Overall, these findings suggest that KRAS gene mutations may be associated with alterations in the intestinal microbiota, highlighting the importance of investigating the role of the microbiota in cancer development and progression.
EGFR mutation-associated airway microbiota
In 2021, a study collected 47 samples of lung microbiota obtained by bronchoalveolar lavage. Among these, there were 32 samples that were obtained from patients diagnosed of EGFR mutated (EGFRm) NSCLC. The study revealed an abundance of Rhizopus oryzae, Natronolimnobius innermongolicus, Staphylococcus sciuri, Orenia marismortui, Burkholderia multivorans and Sinorhizobium in the EGFRm subgroup (60). Huang et al. conducted another study involving 85 patients diagnosed with lung cancer, 66 of which were adenocarcinomas, with 21 being EGFRm. The analysis of sputum microbiota showed an enrichment of phylum Bacteroidetes and Tenericutes, genera Sharpea, Prevotella, Porphyromonas, Parvimonas, Desulfovibrio, Mycoplasma, Actinobacillus, Dialister, and Eikenella in EGFRm patients compared to those with wild-type EGFR. Subgroup analysis of non-smokers showed an abundance of phylum Bacteroidetes and genera Parvimonas and Actinobacillus. Differences were also observed between patients with early-stage and metastatic disease (61). It is worth mentioning that bacteria Parvimonas—specifically P. micra—has been previously associated to CRC due to its role in promoting an inflammatory microenvironment and contributing to carcinogenesis (70). Actinobacillus has also been described as influencing the production of inflammatory cytokines, appearing enriched in COPD, and potentially playing a role in the development of lung carcinogenesis (71). Tenericutes and Bacteroidetes have been reported to exacerbate COPD by contributing to the maintenance of a chronic inflammatory response in bronchial mucosa (72). These results highlight the association between chronic inflammation and a pro-tumorigenic role of certain bacteria present in EGFRm patients, leading to lung carcinogenesis independently of cigarette-smoke damage in lung epithelial tissue.
EGFR mutation-associated gut microbiota
Concerning EGFRm-associated gut microbiota, a study involving 37 female never-smokers diagnosed with lung adenocarcinoma investigated the relationship between cancer progression and gut microbiota, taking into consideration EGFR status as one of the variables (62). The analysis revealed that the presence of EGFR mutation, observed in 56% of the patients, did not show a statistically significant impact on gut microbiota composition. However, it is worth mentioning that Bifidobacterium and Faecalibacterium were more predominant in EGFR wild-type patients compared to mutated patients, whereas Blautia was less abundant in EGFR wild-type patients. Interestingly, previous studies have associated an improved response to immunotherapy with the enrichment of Bifidobacterium, Faecalibacterium, and Akkermansia (73). This suggests a connection between gut microbiota and treatment response, supporting the hypothesis that resistance to ICB in EGFRm patients might be related to microbiota composition.
Saifon et al. compared the gut microbiota of 13 patients with EGFR wild-type and 15 EGFRm NSCLC patients. An abundance of Proteobacteria was found in the EGFR wild-type cohort, while the mutated patients had a predominance of Bacteroidetes and Firmicutes, bacteria that have been previously associated with severe COPD and lung cancer (63,74,75). They also analyzed changes in the microbiota composition after treatment with chemotherapy in the EGFR wild-type group, and after TKI treatment in the EGFRm cohort. However, no statistically significant differences were found. Microbiota did change in response to chemotherapy, with a decrease in Proteobacteria and an increase in Bacteroidetes and Firmicutes. These two last phyla were mostly enriched in patients who experienced severe adverse events. Concerning response rates, Actinobacteria was predominant in EGFRm patients who exhibited progressive disease at first evaluation after treatment with a TKI. Previous evidence has associated Actinobacteria to breast and lung cancer, suggesting a protective role for oral cavity cancer, whereas other studies have linked its presence to increased response to anti-PD-1 therapy in NSCLC patients (76,77). These findings highlight the intricate network between specific bacteria, cancer types, TME, therapy used and immune activity, supporting a multifactorial interaction model.
A recent study assessed gut microbiota composition and ICB efficacy in patients with EGFRm NSCLC. Responders exhibited an enrichment of species Bradyrhizobium guangdongense, Plantactinospora sp. BC1, Corynebacterium stationis, and Methanococcus vannielii. No similarities were found with the microbiota enriched in EGFR wild-type responder patients, which included Akkermansia muciniphila, Bifidobacterium bifidum, or Bifidobacterium breve. Interestingly, the use of antibiotics was correlated with worse outcomes, suggesting a role for dysbiosis and dynamic microbiota changes in treatment efficacy (78).
EGFR mutation-associated intratumoral microbiota
When analyzing EGFR-associated intratumoral microbiota, Zhang et al. published interesting results that correlated EGFR mutation with the intratumoral presence of Serratia marcescens (S. marcescens) in patients with NSCLC (64). S. marcescens is known to produce prodigiosin, a secondary metabolite with cytotoxic and immunosuppressive activity that leads cells to apoptosis using the mitochondrial pathway (79). In this study, this bacterion was associated with better OS, while Haemophilus parainfluenzae was linked to worse prognosis. The presence of EGFR mutation also correlated negatively with Haemophilus parainfluenzae.
Overall, although there is currently insufficient evidence to establish the impact of microbiota in EGFR-driven NSCLC, it represents a field of active research. This might help us understand, in a near future, the role of microbes in the effectiveness of TKIs, the occurrence of TKI-related adverse effects, and their influence on the modulation of the oncogene-addicted TME as well as the immune system. Moreover, it has the potential to provide valuable insights of more effective treatment strategies for oncogene-addicted tumors.
MAPK pathway in other tumors and microbiome
Focusing on the microbiota and its relationship to MAPK pathway in other tumors, a study demonstrated that the presence of Enterococcus faecalis in the oropharynx induced activation of the EGFR pathway in 20 patients with oral cancer, promoting cell proliferation by the production of H2O2 or EGF-like signals such as TNFα (65). In the case of Helicobacter pylori (H. pylori), its presence in gastric tissue has been linked to an increase of EGFR gene expression in the antrum, in order to facilitate the healing of ulcers caused by this pathogen. The EGFR expression levels seemed to return to baseline after successful eradication of H. pylori. The increased EGFR activity induced by epithelial injury promoted cell growth, thus increasing the risk of gastric cancer (66).
A review published by Mihai et al. proposed a potential role of antibiotics in order to manage EGFR inhibitors side effects in CRC, which may eventually also impact prognosis. This is based on the association of Fusobacterium enrichment with poor prognosis and resistance to chemotherapy in CRC patients. Notably, doxycycline, the antibiotic of choice for managing EGFR inhibitor-induced skin rash, exhibits anti-Fusobacterium and anti-anaerobic properties, suggesting a potential benefit for CRC patients (80). Another strategy mentioned in preclinical studies is the inhibition of EGFR pathway by probiotics (81). This might be an interesting approach in molecularly driven EGFR tumors. However, evidence is lacking and definite conclusions cannot be drawn from the available studies.
Her2, another member of the EGFR family, has been extensively studied in breast cancer due to the impact of targeted treatments on the prognosis of patients. Her2-overexpressed breast cancer patients show less microbiota diversity, lower levels of certain Firmicutes—Clostridium, Blautia, Coprococcus, Ruminococcus—and higher levels of Bacteroidetes when compared to Her2-negative patients (82). Microbiota composition might also have an impact on treatment efficacy in this subset of patients. It has been described that diminished levels of Lachnospiraceae, Turicibacteraceae, Bifidobacteriaceae, and Prevotellaceae in gut microbiota were associated to non-responders treated with trastuzumab, an antibody targeting Her2 (83).
TKIs
Microbiota composition has been described to alter drug concentrations by several mechanisms (84). The first-pass effect, by which the drug is taken orally and is metabolized in the gastrointestinal tract, might be influenced by gut microbiota. Microbes can also induce activation of prodrugs, alter the enterohepatic recirculation by secreting β-glucuronidases or affect bile acids composition by microbial bile salt hydrolase (BSH) enzymes and microbial 7α-dehydroxylases. All of these mechanisms influence the absorption, bioavailability and efficacy of oral drugs (85). The fact that TKIs are administered orally reveals the importance of gut microbiota composition and diversity when using these agents, as they might participate in drug metabolism and absorption, influencing pharmacokinetics as well as toxicity (86).
TKI and microbiota
Osimertinib, a third generation EGFR-inhibitor and currently the standard of care in first-line setting for EGFR oncogene-addicted lung cancer, has been described to impact on gut microbiome. However, no significant differences were observed in microbiota composition when comparing the 8 NSCLC EGFRm patients with 21 healthy individuals, nor changes were seen when comparing pre- and post-therapeutic samples (87). As regards erlotinib, a first-generation EGFR-inhibitor, a preclinical study associated the presence of Bacteroides ovatus and Bacteroides xylanisolvens in lung cancer samples treated with this drug with better outcomes. This study also reported an increase in chemokines related to anti-tumor responses, such as CXCL9 and IFN-γ, and increased OS, suggesting a synergistic effect between erlotinib and the microbiota (88-91). A retrospective analysis carried out by Tinsley et al., which included 168 patients diagnosed with advanced melanoma and NSCLC, showed a decrease in progression-free survival (PFS) and OS in patients who were exposed to antibiotics during TKI treatment. The TKIs administered included BRAF inhibitor dabrafenib, MEK inhibitor vemurafenib, and EGFR inhibitors gefitinib, afatinib and erlotinib. The postulated hypothesis was a reduction in number and diversity of microbes due to antibiotics depleting gut microbiota, although this causal link could not be confirmed. The study also emphasized the importance of cytochrome 3A4 inhibition by certain medications, such as macrolides or antifungals, or its induction by rifampicin, which may have altered TKIs blood concentration thus affecting treatment response (92). A recently published preclinical study focused on the impact of microbiota in gefitinib efficacy in lung adenocarcinoma. The presence of genera Prevotellamassilia, Duncaniella, Prevotella, Marinilabilia, and Bacteroides in mice appeared to regulate tumor growth, suggesting a potential role of these microorganisms in the antitumor efficacy of gefitinib (93).
Focusing on other tumors and their respective targeted treatments, a retrospective study was performed in 145 patients with metastatic renal cell carcinoma receiving vascular endothelial growth factor (VEGF) TKIs. Among these patients, those who had received antibiotics covering Bacteroides spp.—beta-lactam antimicrobials, clindamycin, metronidazole, carbapenem—had improved PFS compared with those who received no antibiotics or antibiotics without coverage for Bacteroides spp. (94). Evidence supports this by demonstrating an increase in Bacteroides abundance in response to cytostatic agent-induced diarrhea. Administering antibiotics that reduce these bacteria might enhance both tolerance and effectiveness of TKIs, as diarrhea is a common adverse effect that leads to dose reductions. However, no statistically significant association was found between TKI dose reductions and PFS, suggesting antibiotic administration has an effect on PFS that is independent of diarrhea (95). Further evidence highlights the fact that diarrhea alters microbiota composition by reducing its richness and diversity, and favoring the abundance of species that induce intestinal damage such as C. perfringens, E. coli, Staphylococcus, Bacillus, Enterococcus Acinetobacter, Streptococcus. These species have been previously linked to irinotecan-induced dysbiosis (96,97).
Pomej et al. found worse outcomes in patients diagnosed with hepatocarcinoma treated with sorafenib who received antibiotics due to bacterial infections or hepatic encephalopathy. This further supports the hypothesis that changes in composition of gut microbiota due to antibiotic administration might have an impact on TKI efficacy (98).
TKI, adverse effects and impact on microbiota
A frequent adverse effect of patients under TKIs is diarrhea, which might consequently alter the intestinal environment generating dysbiosis. An example of this was seen by a decrease in microbial diversity in a preclinical study with lapatinib, an EGFR inhibitor used in breast cancer (99). Another study revealed that patients diagnosed with metastatic renal cell carcinoma treated with VEGF TKIs who presented diarrhea had an abundance of Bacteroides spp. and lower levels of Prevotella and Bifidobacterium (95). Secombe et al. highlighted that rash is also a recurrent adverse effect of TKIs which sometimes requires the use of antibiotics to ameliorate it, thus causing a detrimental effect on gut microbiome (100). In fact, a cohort of 102 patients with NSCLC treated with EGFR TKIs was studied, and results suggested antibiotic use could be a negative predictor for efficacy and toxicity of treatment, with worse PFS and an increase in incidence of dyspnea and diarrhea (101). Microbiota might also influence the appearance of adverse effects, as previously described in a study that showed that patients treated with sorafenib for hepatocellular carcinoma had a reduced incidence of hand-and-foot syndrome when an abundance of Veillonella, Bacillus, Enterobacter, Faecalibacterium, Lachnospira, Dialister, and Anaerostipes was evidenced in gut microbiota. In addition, a lower incidence of diarrhea was seen in patients with increased levels of Butyricimonas (102).
As a result, evidence supports the impact of microbiota in TKI absorption and the resulting adverse effects of these drugs, which may influence response to treatment and prognosis (103). This is of particular importance as they are orally administered drugs, so gut microbiota interacts directly with their pharmacokinetics, as has been already described for other drugs, including digoxin, L-Dopa and non-steroidal anti-inflammatory drugs (104). It is also well known the influence of diet on gut microbiome, and the effect of food intake on certain TKIs such as gefitinib, erlotinib and afatinib by increasing the absorption in 37%, 40% and 50%, respectively (103).
To sum up, the evidence that links TKIs and microbiota places special emphasis on the role of diarrhea as a frequent adverse effect of these drugs, as well as the use of antibiotics and dietary habits, which alter microbiota composition and thus impact results to therapy.
Dietary interventions and future directions
The emergence of oncogene-addicted tumors has reshaped our understanding of cancer biology, particularly in non-smokers, where these tumors play an increasingly prominent role. As we delve into the intricacies of these tumors, the role of the microbiota is gaining recognition as a crucial determinant. The relationship between oncogene-addicted tumors and alterations in the microbiota presents a fascinating avenue for future research. Understanding how specific oncogenic drivers interact with the microbiome, and how these interactions influence tumor behavior and treatment response, holds the potential to revolutionize targeted therapies. Exploring the dynamic interplay between oncogenic mutations, the host’s microbiota, and the tumor microenvironment is poised to uncover novel therapeutic strategies. As we navigate this evolving landscape, future directions in cancer research must prioritize unraveling the intricate connections between oncogene addiction and microbiota changes, paving the way for more effective and personalized treatment approaches and lifestyle interventions.
Intervening dietary habits is an appealing approach, particularly supported by the observation that the Mediterranean diet—characterized by high dietary fiber and low processed food intake—has been associated with promoting greater biodiversity and an abundance of beneficial bacteria such as Bacteroides, Lactobacilly, Bifidobacteria or Faecalibacterium, along with a decrease in Firmicutes and Proteobacteria (105). This fosters an anti-inflammatory environment and reduces oxidative stress, maintaining homeostasis (106). Figure 3 summarizes these interactions. Other interventions such as the use of probiotics, prebiotics, synbiotics and FMT appear to be promising strategies. Additionally, it’s crucial to recognize that adverse effects of cancer treatments, such as diarrhea, and the frequent use of antibiotics in immunosuppressive patients, impact microbiome composition and reduce microbial diversity (107,108). As a result, there are many clinical trials ongoing in order to investigate the role of microbiome in response to immunotherapy and dietary interventions. However, there is a noticeable scarcity of trials focusing on oncogenic-driven lung cancer, microbiome interactions, and the impact of targeted therapies. This research gap limits our understanding of the intricate interplay among targeted therapies, lung cancer, and the microbiota (109). Table 3 summarizes the ongoing clinical trials regarding NSCLC treatment and microbiome.
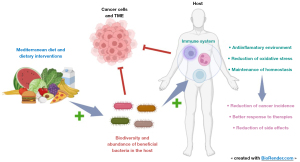
Table 3
| Clinical trial | Study title | Study type | Aims |
|---|---|---|---|
| NCT05502913 | Fecal Microbiota Transplantation With Immune Checkpoint Inhibitors in Lung Cancer | Interventional, recruiting | Safety and efficacy of FMT treatment combined with first-line (chemo-)immunotherapy in metastatic lung cancer |
| NCT04680377 | Using Microbiome to Predict Durvalumab Toxicity in Post- Concurrent Chemoradiation Therapy (CCRT) NSCLC Patients (Microdurva) | Observational, recruiting | Determine if examining the microbiome in non-small cell lung cancer participants who will receive durvalumab can predict treatment toxicity |
| NCT04107168 | Microbiome Immunotherapy Toxicity and Response Evaluation | Observational, recruiting | Saliva and a series of stool samples will be collected from patients with melanoma, renal or lung cancer receiving checkpoint inhibitors to analyse their microbiome and will be linked to treatment response |
| NCT05037825 | The Gut Microbiome and Immune Checkpoint Inhibitor Therapy in Solid Tumors | Observational, recruiting | Assess the associations between the gut microbiota (composition and function), host immune system, and ICI treatment efficacy across multiple cancer types |
| NCT04711330 | Response and Toxicity Prediction by Microbiome Analysis After Concurrent Chemoradiotherapy | Observational, recruiting | The predictive value of the microbiome (throat swabs, stool and of bronchial samples) to identify patients who will relapse during durvalumab treatment after CRT (false negative rate) at 6 months |
| NCT03068663 | Microbiota and the Lung Cancer (MICA) | Interventional, recruiting | Study the composition of the microbiota from the lung, the upper airways and the gut, in patients who undergo surgical treatment or chemotherapy. These results will serve as a base for a future study, focused on manipulation of the microbiota by prebiotics, probiotics or synbiotics |
| NCT05303493 | Camu-Camu Prebiotic and Immune Checkpoint Inhibition in Patients With Non-small Cell Lung Cancer and Melanoma | Interventional, recruiting | Determine safety and tolerability and measure objective response rate in patients treated with checkpoint inhibitors and Camu Camu (prebiotic potential) |
| NCT04965129 | Supplementation of n-3 PUFA in the Modulation of Lean Mass in Patients With Lung Cancer Receiving a High-protein Diet | Interventional, recruiting | The purpose of this study is to assess the effects of fish oil supplementation in the modulation of lean mass and intestinal microbiome in patients with lung cancer undergoing treatment with immunotherapy, chemotherapy and tyrosine kinase inhibitors receiving a high-protein diet |
| NCT04636775 | Microbiome in Immunotherapy naïve NSCLC Patients Receiving PD-1/L1 Blockade (MIP_NSCLC) | Observational, recruiting | Determine if examining the microbiome in non-small cell lung cancer, immunotherapy naive participants can predict the effectiveness of immunotherapy treatment as well as determine ahead of time adverse events and their severity. In addition, the investigator will look into microbiome changing modifiers |
| NCT05037825 | The Gut Microbiome and Immune Checkpoint Inhibitor Therapy in Solid Tumors (PARADIGM) | Observational, recruiting | Large cohort design to assess the associations between the gut microbiota, host immune system, and ICB treatment efficacy across multiple cancer types |
| NCT05777603 | Study of Aerosolized Antibiotics and Pembrolizumab in Advanced Non-small Cell Lung Cancer | Interventional, recruiting | To test two inhaled antibiotics (aztreonam and vancomycin), combined with a standard cancer treatment, in people with NSCLC. Bacterial changes may aid in treatment efficacy |
| NCT05286294 | Microbiota Transplant to Cancer Patients Who Have Failed Immunotherapy Using Faeces From Clinical Responders (MITRIC) | Interventional, recruiting | Phase Iia study evaluating the safety, feasibility and efficacy of FMT to cancer patients not responding to ICB therapy, using ICB-responders as donors |
NSCLC, non-small cell lung cancer; ICI, immune-checkpoint inhibitor; CRT, chemo-radiotherapy; ICB, immune checkpoint blocker; FMT, fecal microbiota transplant.
In summary, these interventions have promising therapeutic potential and represent a challenging strategy to enhance clinical response to oncological treatments with targeted therapies. Changes in microbiome due to dietary habits and lifestyle may help in this setting, as well as in reducing toxicities. Indeed, interventions for general population and environment may help to reduce cancer incidence, as well as side effects due to immunotherapies and targeted therapies for solid tumors. Nevertheless, there are still some uncertainties on the risks they might represent. It is more and more clear that cancer strategies require public interventions beyond oncology approaches. To better address these uncertainties, translational and preclinical research strategies are clearly needed and will provide better evidence for this scenario.
Conclusions
Oncogene-addicted lung cancer prognosis has changed dramatically since the appraisal of TKIs. Emerging evidence suggests that the microbiota may influence TKI effectiveness, due to its impact in drug absorption and metabolism. While extensive research is being conducted on the immunomodulatory role of the microbiota, as well as its impact on response to treatments and the occurrence of adverse effects, particularly with ICB, limited evidence addresses its role in targeted therapies.
Understanding the complex interaction between the microbiota, the immune system and oncogene-driven tumors could unveil prognostic and predictive biomarkers that can enhance the approach of oncogene-addicted lung cancer. New therapeutic strategies involving interventions in the microbiome, such as FMT or the use of probiotics, prebiotics, synbiotics, or changes in dietary habits, offer encouraging pathways to improve outcomes in this challenging and deadly disease. Although much has been learnt about the microbiome, the immune system, and cancer treatments in the past few years, there is still a long pathway to be unraveled.
Acknowledgments
We would like to thank our colleagues at the Medical Oncology Intercenter Unit and especially the patients, without whom treatments and research would not be possible.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-216/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-216/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-216/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li X, Zhang S, Guo G, et al. Gut microbiome in modulating immune checkpoint inhibitors. EBioMedicine 2022;82:104163. [Crossref] [PubMed]
- Zhang C, Wang J, Sun Z, et al. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci 2021;112:3005-17. [Crossref] [PubMed]
- Bredin P, Naidoo J. The gut microbiome, immune check point inhibition and immune-related adverse events in non-small cell lung cancer. Cancer Metastasis Rev 2022;41:347-66. [Crossref] [PubMed]
- Yagi K, Huffnagle GB, Lukacs NW, et al. The Lung Microbiome during Health and Disease. Int J Mol Sci 2021;22:10872. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Martinez FJ, et al. The Microbiome and the Respiratory Tract. Annu Rev Physiol 2016;78:481-504. [Crossref] [PubMed]
- Yu G, Gail MH, Consonni D, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol 2016;17:163. [Crossref] [PubMed]
- Raudoniute J, Bironaite D, Bagdonas E, et al. Human airway and lung microbiome at the crossroad of health and disease Exp Ther Med 2023;25:18. (Review). [Crossref] [PubMed]
- Russo C, Colaianni V, Ielo G, et al. Impact of Lung Microbiota on COPD. Biomedicines 2022;10:1337. [Crossref] [PubMed]
- Mathieu E, Escribano-Vazquez U, Descamps D, et al. Paradigms of Lung Microbiota Functions in Health and Disease, Particularly, in Asthma. Front Physiol 2018;9:1168. [Crossref] [PubMed]
- Dong SR, Han YJ, Wu J, et al. Distribution of Microbiota in Fine Particulate Matter Particles in Guangzhou, China. Biomed Environ Sci 2020;33:306-14. [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Jassem J. Lung cancer: a preventable disease. Transl Lung Cancer Res 2019;8:S1-2. [Crossref] [PubMed]
- Cicchinelli S, Rosa F, Manca F, et al. The Impact of Smoking on Microbiota: A Narrative Review. Biomedicines 2023;11:1144. [Crossref] [PubMed]
- Hou K, Wu ZX, Chen XY, et al. Microbiota in health and diseases. Signal Transduct Target Ther 2022;7:135. [Crossref] [PubMed]
- Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084-9. [Crossref] [PubMed]
- Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079-84. [Crossref] [PubMed]
- Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97-103. [Crossref] [PubMed]
- Alkan Şen G, Şentürk Öztaş N, Değerli E, et al. Effect of antibiotic treatment on immune checkpoint inhibitors efficacy in patients with advanced non-small cell lung cancer. Lung Cancer 2023;184:107347. [Crossref] [PubMed]
- Chen Q, Zhang Z, Li X, et al. Analysis of prognostic factors affecting immune checkpoint inhibitor therapy in tumor patients exposed to antibiotics. Front Oncol 2023;13:1204248. [Crossref] [PubMed]
- Wang B, Yao M, Lv L, et al. The Human Microbiota in Health and Disease. Engineering 2017;3:71-82. [Crossref]
- Yan X, Yang M, Liu J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res 2015;5:3111-22. [PubMed]
- Huang YJ, Sethi S, Murphy T, et al. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol 2014;52:2813-23. [Crossref] [PubMed]
- Mammen MJ, Sethi S. COPD and the microbiome. Respirology 2016;21:590-9. [Crossref] [PubMed]
- Guardamagna M, Berciano-Guerrero MA, Villaescusa-González B, et al. Gut Microbiota and Therapy in Metastatic Melanoma: Focus on MAPK Pathway Inhibition. Int J Mol Sci 2022;23:11990. [Crossref] [PubMed]
- Mekadim C, Skalnikova HK, Cizkova J, et al. Dysbiosis of skin microbiome and gut microbiome in melanoma progression. BMC Microbiol 2022;22:63. [Crossref] [PubMed]
- National Library of Medicine. Pubmed Timeline results Carcinogenesis AND Microbiome. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=Carcinogénesis%20AND%20microbiome&timeline=expanded (2023).
- Knippel RJ, Drewes JL, Sears CL. The Cancer Microbiome: Recent Highlights and Knowledge Gaps. Cancer Discov 2021;11:2378-95. [Crossref] [PubMed]
- Ternes D, Karta J, Tsenkova M, et al. Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism? Trends Microbiol 2020;28:401-23. [Crossref] [PubMed]
- Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019;51:27-41. [Crossref] [PubMed]
- Takahashi-Kanemitsu A, Knight CT, Hatakeyama M. Molecular anatomy and pathogenic actions of Helicobacter pylori CagA that underpin gastric carcinogenesis. Cell Mol Immunol 2020;17:50-63. [Crossref] [PubMed]
- Elagan SK, Almalki SJ, Alharthi MR, et al. Role of Bacteria in the Incidence of Common GIT Cancers: The Dialectical Role of Integrated Bacterial DNA in Human Carcinogenesis. Infect Drug Resist 2021;14:2003-14. [Crossref] [PubMed]
- Pérez-Ruiz E, Jiménez-Castro J, Berciano-Guerrero MA, et al. Impact of intestinal dysbiosis-related drugs on the efficacy of immune checkpoint inhibitors in clinical practice. Clin Transl Oncol 2020;22:1778-85. [Crossref] [PubMed]
- Roviello G, Iannone LF, Bersanelli M, et al. The gut microbiome and efficacy of cancer immunotherapy. Pharmacol Ther 2022;231:107973. [Crossref] [PubMed]
- Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012;3:4-14. [Crossref] [PubMed]
- Owaga E, Hsieh RH, Mugendi B, et al. Th17 Cells as Potential Probiotic Therapeutic Targets in Inflammatory Bowel Diseases. Int J Mol Sci 2015;16:20841-58. [Crossref] [PubMed]
- Bhat T, Teli S, Rijal J, et al. Neutrophil to lymphocyte ratio and cardiovascular diseases: a review. Expert Rev Cardiovasc Ther 2013;11:55-9. [Crossref] [PubMed]
- Langley BO, Guedry SE, Goldenberg JZ, et al. Inflammatory Bowel Disease and Neutrophil-Lymphocyte Ratio: A Systematic Scoping Review. J Clin Med 2021;10:4219. [Crossref] [PubMed]
- Howard R, Kanetsky PA, Egan KM. Exploring the prognostic value of the neutrophil-to-lymphocyte ratio in cancer. Sci Rep 2019;9:19673. [Crossref] [PubMed]
- Yoon HY, Kim HN, Lee SH, et al. Association between Neutrophil-to-Lymphocyte Ratio and Gut Microbiota in a Large Population: a Retrospective Cross-Sectional Study. Sci Rep 2018;8:16031. [Crossref] [PubMed]
- Valero C, Lee M, Hoen D, et al. Pretreatment neutrophil-to-lymphocyte ratio and mutational burden as biomarkers of tumor response to immune checkpoint inhibitors. Nat Commun 2021;12:729. [Crossref] [PubMed]
- Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res 2020;30:492-506. [Crossref] [PubMed]
- Anderson NM, Simon MC. The tumor microenvironment. Curr Biol 2020;30:R921-5. [Crossref] [PubMed]
- Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol 2014;16:1024-33. [Crossref] [PubMed]
- Murphy JF. The human microbiome and the tumor microenvironment. Explor Immunol 2022;2:581-8. [Crossref]
- Jiang X, Wang J, Deng X, et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res 2020;39:204. [Crossref] [PubMed]
- He Y, Huang J, Li Q, et al. Gut Microbiota and Tumor Immune Escape: A New Perspective for Improving Tumor Immunotherapy. Cancers (Basel) 2022;14:5317. [Crossref] [PubMed]
- Boesch M, Horvath L, Baty F, et al. Compartmentalization of the host microbiome: how tumor microbiota shapes checkpoint immunotherapy outcome and offers therapeutic prospects. J Immunother Cancer 2022;10:e005401. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: An overview. Int J Cancer 2021; Epub ahead of print. [Crossref] [PubMed]
- Gomes S, Cavadas B, Ferreira JC, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci Rep 2019;9:12838. [Crossref] [PubMed]
- Ramírez-Labrada AG, Isla D, Artal A, et al. The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy. Trends Cancer 2020;6:86-97. [Crossref] [PubMed]
- Greathouse KL, White JR, Vargas AJ, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol 2018;19:123. [Crossref] [PubMed]
- Zhou Y, Chen T. Human microbiota: A crucial gatekeeper in lung cancer initiation, progression, and treatment. Med Microecol 2022;13:100055. [Crossref]
- Liu X, Cheng Y, Zang D, et al. The Role of Gut Microbiota in Lung Cancer: From Carcinogenesis to Immunotherapy. Front Oncol 2021;11:720842. [Crossref] [PubMed]
- Peters BA, Hayes RB, Goparaju C, et al. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol Biomarkers Prev 2019;28:731-40. [Crossref] [PubMed]
- Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368:973-80. [Crossref] [PubMed]
- Saw SPL, Le X, Hendriks LEL, et al. New Treatment Options for Patients With Oncogene-Addicted Non-Small Cell Lung Cancer Focusing on EGFR-Mutant Tumors. Am Soc Clin Oncol Educ Book 2024;44:e432516. [Crossref] [PubMed]
- Zhu QG, Zhang SM, Ding XX, et al. Driver genes in non-small cell lung cancer: Characteristics, detection methods, and targeted therapies. Oncotarget 2017;8:57680-92. [Crossref] [PubMed]
- Black RC, Khurshid H. NSCLC: An Update of Driver Mutations, Their Role in Pathogenesis and Clinical Significance. R I Med J (2013) 2015;98:25-8. [PubMed]
- Jin C, Lagoudas GK, Zhao C, et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019;176:998-1013.e16. [Crossref] [PubMed]
- Zheng L, Sun R, Zhu Y, et al. Lung microbiome alterations in NSCLC patients. Sci Rep 2021;11:11736. [Crossref] [PubMed]
- Huang DH, He J, Su XF, et al. The airway microbiota of non-small cell lung cancer patients and its relationship to tumor stage and EGFR gene mutation. Thorac Cancer 2022;13:858-69. [Crossref] [PubMed]
- Otoshi T, Nagano T, Park J, et al. The Gut Microbiome as a Biomarker of Cancer Progression Among Female Never-smokers With Lung Adenocarcinoma. Anticancer Res 2022;42:1589-98. [Crossref] [PubMed]
- Saifon W, Sensorn I, Trachu N, et al. Gastrointestinal microbiota profile and clinical correlations in advanced EGFR-WT and EGFR-mutant non-small cell lung cancer. BMC Cancer 2022;22:963. [Crossref] [PubMed]
- Zhang M, Zhang Y, Sun Y, et al. Intratumoral Microbiota Impacts the First-Line Treatment Efficacy and Survival in Non-Small Cell Lung Cancer Patients Free of Lung Infection. J Healthc Eng 2022;2022:5466853. [Crossref] [PubMed]
- Boonanantanasarn K, Gill AL, Yap Y, et al. Enterococcus faecalis enhances cell proliferation through hydrogen peroxide-mediated epidermal growth factor receptor activation. Infect Immun 2012;80:3545-58. [Crossref] [PubMed]
- Wong BC, Wang WP, So WH, et al. Epidermal growth factor and its receptor in chronic active gastritis and gastroduodenal ulcer before and after Helicobacter pylori eradication. Aliment Pharmacol Ther 2001;15:1459-65. [Crossref] [PubMed]
- Kirchberger S, Royston DJ, Boulard O, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med 2013;210:917-31. [Crossref] [PubMed]
- Sui X, Chen Y, Liu B, et al. The relationship between KRAS gene mutation and intestinal flora in tumor tissues of colorectal cancer patients. Ann Transl Med 2020;8:1085. [Crossref] [PubMed]
- Kim J, Lee HK. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front Immunol 2021;12:807648. [Crossref] [PubMed]
- Xu J, Yang M, Wang D, et al. Alteration of the abundance of Parvimonas micra in the gut along the adenoma-carcinoma sequence. Oncol Lett 2020;20:106. [Crossref] [PubMed]
- Haldar K, George L, Wang Z, et al. The sputum microbiome is distinct between COPD and health, independent of smoking history. Respir Res 2020;21:183. [Crossref] [PubMed]
- Garcia-Nuñez M, Millares L, Pomares X, et al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol 2014;52:4217-23. [Crossref] [PubMed]
- Wu X, Zhang T, Chen X, et al. Microbiota transplantation: Targeting cancer treatment. Cancer Lett 2019;452:144-51. [Crossref] [PubMed]
- Lee SH, Sung JY, Yong D, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016;102:89-95. [Crossref] [PubMed]
- Sze MA, Dimitriu PA, Hayashi S, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2012;185:1073-80. [Crossref] [PubMed]
- Long Y, Tang L, Zhou Y, et al. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med 2023;21:66. [Crossref] [PubMed]
- Song P, Yang D, Wang H, et al. Relationship between intestinal flora structure and metabolite analysis and immunotherapy efficacy in Chinese NSCLC patients. Thorac Cancer 2020;11:1621-32. [Crossref] [PubMed]
- Luo WC, Mei SQ, Huang ZJ, et al. Correlation of distribution characteristics and dynamic changes of gut microbiota with the efficacy of immunotherapy in EGFR-mutated non-small cell lung cancer. J Transl Med 2024;22:326. [Crossref] [PubMed]
- Anwar MM, Albanese C, Hamdy NM, et al. Rise of the natural red pigment 'prodigiosin' as an immunomodulator in cancer. Cancer Cell Int 2022;22:419. [Crossref] [PubMed]
- Mihai MM, Ion A, Giurcăneanu C, et al. The Impact of Long-Term Antibiotic Therapy of Cutaneous Adverse Reactions to EGFR Inhibitors in Colorectal Cancer Patients. J Clin Med 2021;10:3219. [Crossref] [PubMed]
- Ma EL, Choi YJ, Choi J, et al. The anticancer effect of probiotic Bacillus polyfermenticus on human colon cancer cells is mediated through ErbB2 and ErbB3 inhibition. Int J Cancer 2010;127:780-90. [Crossref] [PubMed]
- Di Modica M, Arlotta V, Sfondrini L, et al. The Link Between the Microbiota and HER2+ Breast Cancer: The New Challenge of Precision Medicine. Front Oncol 2022;12:947188. [Crossref] [PubMed]
- Di Modica M, Gargari G, Regondi V, et al. Gut Microbiota Condition the Therapeutic Efficacy of Trastuzumab in HER2-Positive Breast Cancer. Cancer Res 2021;81:2195-206. [Crossref] [PubMed]
- Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res 2017;179:204-22. [Crossref] [PubMed]
- Zhang X, Han Y, Huang W, et al. The influence of the gut microbiota on the bioavailability of oral drugs. Acta Pharm Sin B 2021;11:1789-812. [Crossref] [PubMed]
- Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, et al. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 2019;363:eaat9931. [Crossref] [PubMed]
- Cong J, Zhang Y, Xue Y, et al. A Pilot Study: Changes of Intestinal Microbiota of Patients With Non-small Cell Lung Cancer in Response to Osimertinib Therapy. Front Microbiol 2020;11:583525. [Crossref] [PubMed]
- Heshiki Y, Vazquez-Uribe R, Li J, et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 2020;8:28. [Crossref] [PubMed]
- Bronger H, Singer J, Windmüller C, et al. CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br J Cancer 2016;115:553-63. [Crossref] [PubMed]
- Jorgovanovic D, Song M, Wang L, et al. Roles of IFN-γ in tumor progression and regression: a review. Biomark Res 2020;8:49. [Crossref] [PubMed]
- Conti G, D'Amico F, Fabbrini M, et al. Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out. Genes (Basel) 2022;14:55. [Crossref] [PubMed]
- Tinsley N, Zhou C, Nahm S, et al. Antibiotic use reduces efficacy of tyrosine kinase inhibitors in patients with advanced melanoma and non-small-cell lung cancer. ESMO Open 2022;7:100430. [Crossref] [PubMed]
- Jiang T, Zhang M, Hao S, et al. Revealing the role of the gut microbiota in enhancing targeted therapy efficacy for lung adenocarcinoma. Exp Hematol Oncol 2024;13:15. [Crossref] [PubMed]
- Hahn AW, Froerer C, VanAlstine S, et al. Targeting Bacteroides in Stool Microbiome and Response to Treatment With First-Line VEGF Tyrosine Kinase Inhibitors in Metastatic Renal-Cell Carcinoma. Clin Genitourin Cancer 2018;16:365-8. [Crossref] [PubMed]
- Pal SK, Li SM, Wu X, et al. Stool Bacteriomic Profiling in Patients with Metastatic Renal Cell Carcinoma Receiving Vascular Endothelial Growth Factor-Tyrosine Kinase Inhibitors. Clin Cancer Res 2015;21:5286-93. [Crossref] [PubMed]
- Chung The H. Le SH. Dynamic of the human gut microbiome under infectious diarrhea. Curr Opin Microbiol 2022;66:79-85. [Crossref] [PubMed]
- Ramamurthy T, Kumari S, Ghosh A. Diarrheal disease and gut microbiome. Prog Mol Biol Transl Sci 2022;192:149-77. [Crossref] [PubMed]
- Pomej K, Balcar L, Scheiner B, et al. Antibiotic Therapy is Associated with Worse Outcome in Patients with Hepatocellular Carcinoma Treated with Sorafenib. J Hepatocell Carcinoma 2021;8:1485-93. [Crossref] [PubMed]
- Mayo BJ, Secombe KR, Wignall AD, et al. The GLP-2 analogue elsiglutide reduces diarrhoea caused by the tyrosine kinase inhibitor lapatinib in rats. Cancer Chemother Pharmacol 2020;85:793-803. [Crossref] [PubMed]
- Secombe KR, Van Sebille YZA, Mayo BJ, et al. Diarrhea Induced by Small Molecule Tyrosine Kinase Inhibitors Compared With Chemotherapy: Potential Role of the Microbiome. Integr Cancer Ther 2020;19:1534735420928493. [Crossref] [PubMed]
- Liu K, Zhang W, Tan Q, et al. Antibiotic use is a negative predictor of the efficacy and toxicity of epidermal growth factor receptor-targeted therapy in advanced non-small cell lung cancer. Oncol Lett 2019;18:2677-83. [Crossref] [PubMed]
- Yamamoto K, Kuzuya T, Honda T, et al. Relationship Between Adverse Events and Microbiomes in Advanced Hepatocellular Carcinoma Patients Treated With Sorafenib. Anticancer Res 2020;40:665-76. [Crossref] [PubMed]
- Honeywell RJ, Kathmann I, Giovannetti E, et al. Epithelial Transfer of the Tyrosine Kinase Inhibitors Erlotinib, Gefitinib, Afatinib, Crizotinib, Sorafenib, Sunitinib, and Dasatinib: Implications for Clinical Resistance. Cancers (Basel) 2020;12:3322. [Crossref] [PubMed]
- Tsunoda SM, Gonzales C, Jarmusch AK, et al. Contribution of the Gut Microbiome to Drug Disposition, Pharmacokinetic and Pharmacodynamic Variability. Clin Pharmacokinet 2021;60:971-84. [Crossref] [PubMed]
- Merra G, Noce A, Marrone G, et al. Influence of Mediterranean Diet on Human Gut Microbiota. Nutrients 2020;13:7. [Crossref] [PubMed]
- Nagpal R, Shively CA, Register TC, et al. Gut microbiome-Mediterranean diet interactions in improving host health. F1000Res 2019;8:699. [Crossref] [PubMed]
- Patangia DV, Anthony Ryan C, Dempsey E, et al. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022;11:e1260. [Crossref] [PubMed]
- Gao Y, Shang Q, Li W, et al. Antibiotics for cancer treatment: A double-edged sword. J Cancer 2020;11:5135-49. [Crossref] [PubMed]
- US National Library of Medicine. ClinicalTrials.gov. [Accessed on 30th May 2024]. Available online: https://clinicaltrials.gov/ct2/home


