
Clinical features and prognostic biomarkers in patients with SMARCA4-mutated non-small cell lung cancer
Highlight box
Key findings
• Patients with non-small cell lung cancer (NSCLC) with the SMARCA4 mutation (SMARCA4-Mut) had a worse clinical prognosis than those with wild-type SMARCA4. Napsin A-positive expression in SMARCA4-Mut NSCLC was significantly associated with longer overall survival (OS).
What is known and what is new?
• Patients with NSCLC and SMARCA4-Mut are known to have a worse prognosis than those with the wild-type gene.
• Napsin A expression in SMARCA4-Mut patients is a favorable prognostic factor.
What is the implication, and what should change now?
• NSCLC patients with SMARCA4-Mut should be considered as a distinct entity.
Introduction
According to the 2022 estimates of the Global Cancer Observatory (GLOBOCAN), there were approximately 20 million new cases and 9.7 million cancer deaths worldwide, with lung cancer being the first most common cancer and the leading cause of cancer death (1). Non-small cell lung cancer (NSCLC), which accounts for 85% of lung cancer cases, is associated with low overall survival (OS) and high mortality (2). However, the prognosis of lung cancer varies according to the pathological type and molecular features (3,4).
SMARCA4 gene deletion is a special type of mutation typical for undifferentiated or poorly differentiated NSCLC and is associated with a high grade of malignancy (5). The SMARCA4 gene, located in 19p13.2, encodes the protein BRG1, possesses ATPase activity, and is an important member of the BRG-/BRM-associated factor (BAF) chromosome regulatory complex regulating essential cell biological functions, such as DNA replication and repair, cell division, and differentiation (6). SMARCA4-mutated (SMARCA4-Mut) NSCLC was defined as a new pathological type of NSCLC in the 2021 edition of The WHO Classification of Thoracic Tumors.
SMARCA4 mutations (SMARCA4-Mut) have been reported to occur in about 10% of NSCLC cases, often in conjunction with other well-known lung cancer mutations such as those of KRAS and TP53 (7,8). A study has shown that the median survival time of both SMARCA4-deficient undifferentiated tumors and SMARCA4-Mut non-small cell lung cancer (NSCLC-SMARCA4-Mut) is not ideal, with the former being only 5–7 months (9).
The clinical characteristics of SMARCA4-Mut NSCLC still need to be clarified due to the rarity of this population. We thus aimed to analyze the clinical characteristics and prognosis of patients with SMARCA4-Mut NSCLC by examining cases from The Cancer Genome Atlas (TCGA) database and clinical cases from the 900th Hospital of the Joint Logistic Support Force, People’s Liberation Army of China. Additionally, in the latter group, we investigated the prognostic impact of aspartic peptidase (Napsin A), a crucial biomarker for classifying advanced NSCLC. At present there is no conclusive evidence about the association between SMARCA4 and Napsin A. We combined the latest research reports and the content of this study to try to find new evidence of association between the two. We present this article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-381/rc).
Methods
Patients
Between December 2020 and July 2022, we collected transcriptome and clinical data on NSCLC cancer cases with SMARCA4-Mut from the TCGA database (https://portal.gdc.cancer.gov/) to evaluate the clinical characteristics of SMARCA4-Mut NSCLC patients. The clinical baseline characteristics in this cohort included age, sex, smoking status, pathology, stage, tumor location, previous malignancy history, and treatment. Additionally, we retrospectively identified in the 900th Hospital of the Joint Logistic Support Force, People’s Liberation Army of China database SMARCA4-Mut NSCLC cases to evaluate their clinical, pathological, and molecular features and to assess the prognostic value of SMARCA4-Mut (Figure 1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Committee of the 900th Hospital of the Joint Logistic Support Force, People’s Liberation Army of China (approval No. 2021-026). Informed consent was obtained from all individual participants included in the study. The inclusion criteria were as follows: (I) patients aged ≥18 years with histologically proven NSCLC; (II) patients subjected to next-generation sequencing (NGS) to detect 448 genes or 116 genes, encompassing tissue, blood, and pleural effusion; (III) treated according to the National Comprehensive Cancer Network (NCCN) guidelines.

The collected data included age, sex, smoking history, gene mutations, tissue type, imaging data [primarily computed tomography (CT) and magnetic resonance imaging (MRI) scans], immunohistochemistry (tissue specimen), tumor stage (based on the 8th TNM edition for lung cancer), and treatment. The primary treatment outcomes were OS and first-line treatment progression-free survival (PFS1). OS was defined as the time from the date of diagnosis to the last follow-up and/or death from any causes, while PFS1 was defined as the time from the administration of first-line drug therapy to the date when the first progressive disease (PD) was confirmed. The data cut-off date was on April 30, 2023.
Biopsy tissue specimens were fixed, embedded, sectioned, antigenically repaired, closed, incubated, enzyme-substrate chromogenic or fluorescence detected, and sealed. Additionally, we determined the Napsin A expression using two categories: negative (Napsin A not detected) and positive (Napsin A detected). The study was retrospective and not blinded.
Bioinformatic analysis
Second-generation DNA sequencing was performed with a 448-gene panel and NextSeq 500 system (Illumina Inc., San Diego, CA, USA), which is primarily used for mutation diagnosis in lung and bowel cancers. Finally, bioinformatics and sequencing data were analyzed using the ADXLC10 module of the AmoyDx NGS data analysis system (Amoy Diagnostics Amoy Diagnostics Co., Ltd., Shanghai, China).
Statistical analysis
Statistical analysis was conducted using SPSS Statistics 26.0 software (IBM Corp.). Chi-squared tests or Fisher exact tests were employed to compare the categorical data between the groups. Survival estimates were calculated using the Kaplan-Meier method and compared using the log-rank test. Time-dependent receiver operating characteristic (ROC) curve analysis was used to evaluate the prognostic value of the indicators, and the Cox proportional hazard regression model was used to determine the prognostic factors for OS. All statistical tests were two-sided, and a P value <0.05 was considered statistically significant. GraphPad Prism 9.5.0 (GraphPad Software) was used to generate the survival curve and ROC curve.
Results
Clinicopathological characteristics of the TCGA cohort
A total of 496 cases of SMARCA4-Mut were initially selected from the TCGA database. Of these, 16 cases with incomplete clinical data were excluded, resulting in a final cohort of 480 cases.
The median age in the SMARCA4-Mut TCGA cohort was 67 years (Table 1). A total of 311 patients (64.8%) were males, and 374 (77.9%) were smokers. Among them, 248 (51.7%) were adenocarcinomas and 230 (47.9%) were squamous cell carcinomas. In 277 cases (57.7%) the primary tumor was located in the upper lobe, and in 157 (32.7%) in the lower lobe of the lung.
Table 1
| Characteristics | SMARCA4 mutation group (N=480) |
|---|---|
| Age (years) | |
| ≥18−<60 | 112 (23.3) |
| ≥60−<75 | 282 (58.8) |
| ≥75−<85 | 81 (16.9) |
| ≥85 | 5 (1.0) |
| Gender | |
| Male | 311 (64.8) |
| Female | 169 (35.2) |
| Smoking status | |
| Smoker | 374 (77.9) |
| Never smoker | 106 (22.1) |
| Histology | |
| Squamous cell carcinoma | 230 (47.9) |
| Adenocarcinoma | 248 (51.7) |
| Other† | 2 (0.4) |
| Stages‡ | |
| IA | 104 (21.7) |
| IB | 137 (28.5) |
| IIA | 61 (12.7) |
| IIB | 82 (17.1) |
| IIIA | 63 (13.1) |
| IIIB | 15 (3.1) |
| IV | 18 (3.8) |
| Primary tumor site | |
| Upper lobe | 277 (57.7) |
| Lower lobe | 157 (32.7) |
| Lung, not specified | 21 (4.4) |
| Middle lobe | 14 (3.0) |
| Main bronchus | 5 (1.0) |
| Overlapping lesion | 5 (1.0) |
| Pleura | 1 (0.2) |
| Prior malignancy | |
| Yes | 82 (17.1) |
| No | 398 (82.9) |
| Treatment type | |
| Pharmaceutical | 256 (53.3) |
| Radiation | 224 (46.7) |
Data are presented as n (%). †, small cell, signet ring cell carcinoma, not otherwise specified; ‡, TNM staging of lung cancer (8th edition). TCGA, The Cancer Genome Atlas.
Clinicopathological characteristics of clinical cohort
In our retrospective clinical series, 26 of 224 patients had SMARCA4-Mut and 20 (12 males and 8 females) were eligible for this study, with a median age of 63 years. The control group included 40 NSCLC patients with wild-type SMARCA4 (SMARCA4-WT) diagnosed and treated at our institution during the same period, matched using the propensity score method (1:2 ratio), matching variables including sex, age, smoking status, and pathology type, to ensure that the baseline data are consistent between the two groups.
Hence, a total of 60 patients were analyzed as the clinical cohort. There were no significant differences between both groups in terms of age, sex, smoking status, tumor stage, lymph node metastasis, Histology, or pleural effusion (Table 2).
Table 2
| Characteristics | SMARCA4-Mut group (N=20), n (%) | SMARCA4-WT group (N=40), n (%) | P value |
|---|---|---|---|
| Age (years) | 0.46 | ||
| <65 | 11 (55.0) | 18 (45.0) | |
| ≥65 | 9 (45.0) | 22 (55.0) | |
| Average [range] | 60.15 [34–76] | 61.38 [30–85] | |
| Gender | 0.71 | ||
| Male | 12 (60.0) | 26 (65.0) | |
| Female | 8 (40.0) | 14 (35.0) | |
| Smoking status | 0.55 | ||
| Smoker | 7 (35.0) | 11 (27.5) | |
| Never | 13 (65.0) | 29 (72.5) | |
| Lymph node metastasis | >0.99 | ||
| Yes | 16 (80.0) | 32 (80.0) | |
| No | 4 (20.0) | 8 (20.0) | |
| Histology | 0.20 | ||
| Adenocarcinoma | 14 (70.0) | 32 (80.0) | |
| Squamous cell carcinoma | 2 (10.0) | 6 (15.0) | |
| Other† | 4 (20.0) | 2 (5.0) | |
| Stage | 0.21 | ||
| I–III | 3 (15.0) | 12 (30.0) | |
| IV | 17 (85.0) | 28 (70.0) | |
| Pleural effusion | 0.35 | ||
| Yes | 6 (30.0) | 11 (27.5) | |
| No | 14 (70.0) | 29 (72.5) | |
| EGFR | 0.85 | ||
| Non-mutated | 8 (40.0) | 17 (42.5) | |
| Mutated | 12 (60.0) | 23 (57.5) |
†, large cell lung carcinoma with neuroendocrine features, SMARCA4 undifferentiated lung cancer, undifferentiated lung cancer, sarcomatoid lung carcinoma (one case each). SMARCA4-Mut, SMARCA4 mutation; SMARCA4-WT, SMARCA4 wild type; EGFR, epidermal growth factor receptor.
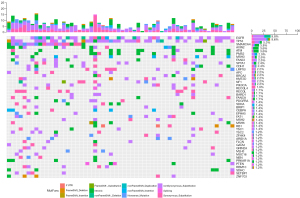
The most common mutations in both groups were in EGFR, TP53, AXIN2, SPTA1, and CDH1 genes. SETBP1 and SPEN were more frequently mutated in the SMARCA4-Mut than in the SMARCA4-WT group (P=0.03) (Figure 2). In the SMARCA4-Mut nonSynonymous_Substitution 7 cases, Synonymous_Substitution 5 cases, intronic 4 cases, Nonsense_Mutation 3 cases, Splicing 1 case. OncoGrid was used to illustrate the frequencies, including a composite of all alterations (5' untranslated region, frameshift mutation, intronic, non-frameshift mutation, nonsense mutation, splicing, and synonymous substitution) (Figure 3).

Prognostic value of SMARCA4-Mut
We excluded from the analysis three cases with no treatment and no survival data in the SMARCA4-Mut group. Thus, prognostic correlation analysis was performed in 17 cases with SMARCA4-Mut and 40 with SMARCA4-WT.
Kaplan-Meier curves were used to analyze whether there was a difference in prognosis between the SMARCA4-Mut and SMARCA4-WT groups. There was a significant difference between the groups in terms of OS (P=0.04) (Figure 4). Consequently, we analyzed the overall samples OS using the Cox proportional risk model. The risk of death in the univariate analysis was statistically higher in the SMARCA4-Mut group (P=0.046) and was in the multivariate analysis (P=0.03) (Figure 5). This suggests that SMARCA4-Mut increases patients’ risk of death, significantly impacting their OS.


Prognostic value of Napsin A expression according to SMARCA4-Mut
In this series, Napsin A expression was less common in the SMARCA4-Mut compared to the SMARCA4-WT NSCLC group (58.8% vs. 65%, respectively), but the difference was insignificant (Table 3).
Table 3
| Characteristics | SMARCA4-Mut group (N=17), n (%) | SMARCA4-WT group (N=40), n (%) | P value |
|---|---|---|---|
| Napsin A | 0.66 | ||
| Negative | 7 (41.2) | 14 (35.0) | |
| Positive | 10 (58.8) | 26 (65.0) | |
| First-line treatment | 0.95 | ||
| Targeted therapy | 9 (52.9) | 23 (57.5) | |
| Chemotherapy + immunotherapy | 6 (35.3) | 13 (32.5) | |
| Nonpharmacological treatments | 2 (11.8) | 4 (10.0) |
SMARCA4-Mut, SMARCA4 mutation; SMARCA4-WT, SMARCA4 wild type.
In the entire group, Napsin A expression was not significantly associated with PFS1 (P=0.63) and OS (P=0.49) (Figure 6). Therefore, we conducted an analysis of the SMARCA4-Mut group and SMARCA4-WT group. There was a statistically significant in OS (P=0.03) between Napsin A-positive and negative patients in the SMARCA4-Mut group (Figure 7). We thus speculated that Napsin A might play a more critical role in the prognosis of SMARCA4-Mut NSCLC.


To further explore the prognostic value of Napsin A, we used time-dependent receiver operating characteristic (time-dependent ROC) curves to predict the impact of Napsin A expression in the SMARCA4-Mut and the SMARCA4-WT groups. In the SMARCA4-Mut group, the area under the curve (AUC) value for PFS1 and OS were 0.748 and 0.586, respectively, and in the SMARCA4-WT group, 0.360 and 0.152, respectively, further supporting the hypothesis of favorable prognostic impact Napsin A expression in SMARCA4-Mut patients (Figure 8).

Discussion
The clinical features of SMARCA4-Mut patients in our clinical sample were similar to those in the TCGA cohort and previous series, considering age, sex, and smoking status. A previous study has indicated that thoracic SMARCA4-deficient undifferentiated tumors are most likely to occur in adults, with a median age of 48 years (ranging from 27 to 90 years), and are significantly more common in males, particularly heavy smokers (10). SMARCA4-deficient NSCLC, a smoking-related undifferentiated or differentiated invasive lung cancer, is predominantly observed in young men with a median age of 63 years and is associated with a high incidence of pleural and vascular infiltration (11).
SMARCA4-Mut NSCLC has been shown to have increased abundances of lymphocytes, macrophages, and multinucleated giant cells, corresponding to an increase in inflammatory cells in the surrounding tumor stroma. The Switch defective/sucrose non-fermentable (SWI/SNF) complex, to which SMARCA4 belongs, has been identified as an immune system modulator. A study has demonstrated that SMARCA4 is a core gene that critically contributes to tumorigenesis by regulating the tumor microenvironment (TME) through both cell autonomy and TME interactions (12). Patients with SMARCA4-Mut NSCLC have a poor prognosis, especially those with truncated, fusion, and homozygous deletion. In turn, SMARCA4-Mut NSCLC, particularly with the last mutation type, may be more sensitive to immune checkpoint inhibitors (ICIs) (7). Several cases of successful ICI treatment for advanced SMARCA4-deficient NSCLC and SMARCA4-deficient malignant rhabdomyoma-like tumors have been reported (8,13).
In our series, SMARCA4-Mut, compared with SMARCA4-WT tumors, more frequently carried mutations in two genes (SETBP1, SPEN), associated with sensitivity to ICI. The SETBP1 gene, present in approximately 13% of patients with lung adenocarcinomas (LUAD), is one of the signatures LUAD mutations, alongside that of KEAP1 and STK11, and carries an adverse prognosis. Low-level expression of SETBP1 induces epithelial-mesenchymal transition (EMT) through the ERK1/2 signaling pathway, promoting proliferation, migration, and invasion of NSCLC cells. SETBP1 expression is significantly correlated with the regulation of tumor-infiltrating immune cells, especially M1 macrophages (14). M1 macrophages are associated with a poor prognosis for lung cancer (15). This study also found that CD1+T cells, CD, and monocytes were negatively correlated with SETBP1 expression levels. SPEN is a tumor-suppressor gene, and its mutations are an adverse prognostic feature in small-cell lung cancer (16). However, SPEN mutation was associated with better OS in a pan-cancer cohort of patients administered ICI (17).
Another finding of our study is a favorable prognostic impact of Napsin A expression in SMARCA4-Mut patients. Napsin A, belonging to the peptidase A1 family, is a single-chain protein with a molecular weight of 45 kDa, composed of 420 amino acids encoded by the NSPSA gene on chromosomal body 19q13.3.45. In the lung, Napsin A is primarily expressed in alveolar type II epithelial cells and in alveolar macrophages. Napsin A is an important biological indicator in typing of advanced NSCLC and a more sensitive marker of LUAD. The loss of SMARCA4 plays a paradoxical role in tumor development, inhabiting tumor progression in early tumors and accelerating at the highly advanced stages (18,19). The reason for this is that most transformed cells carrying SMARCA4-Mut are alveolar type II (ATII) cells, which gradually transform into club cells at the later stage of tumor development and are sensitive to malignant transformation and tumor progression in a cell type-dependent manner (20). We hypothesized that the effect of Napsin A may be related to the transformation of ATII cells in the cancer occurrence and development. High expression of Napsin A predicted a higher proportion of SMARCA4-Mut ATII cells, which in turn was associated with slower tumor progression, leading to prolonged survival.
For the first time, we analyzed the prognostic impact of Napsin A expression in relation to SMARCA4-Mut. Napsin A, belonging to the peptidase A1 family, is a single-chain protein with a molecular weight of 45 kDa, composed of 420 amino acids encoded by the NSPSA gene on chromosomal body 19q13.3.45. In the lung, Napsin A is primarily expressed in alveolar type II epithelial cells and is also present in alveolar macrophages. Napsin A is an important biological indicator in the typing of advanced NSCLC. This protein is mainly expressed in LUAD, whereas SMARCA4-Mut occurs in all pathological types of NSCLC. Napsin A positive expression is associated with a lower grade of malignancy and slower tumor progression (21,22).
Our results indicate that the expression of Napsin A correlates with a better prognosis in patients with SMARCA4-Mut but not in those with SMARCA4-WT. This may also refer to the pathological type.
SMARCA4 deficiency leads to decreased expression of inositol 1,4,5-triphosphate receptor (IP3R3), resulting in impaired Ca2+ transfer from the endoplasmic reticulum to the mitochondria, required for apoptosis induction (23). In turn, SWI/SNF complexes regulate nutrient sensing and energy metabolism during normal development (24). The GLUT1/SLC38A2-mediated metabolic shift may be due to SMARCA4/2 loss. GLUT1 deficiency induced by SMARCA4/2 loss is a key contributor to the oxidative phosphorylation dependency in the SMARCA4/2-deficient cancer cells, and SMARCA4/2-deficient NSCLC cells express the lowest levels of GLUT1 (25). As discussed earlier, the ICI benefits associated with various mutations, and the expression of key cell groups and metabolic components in the TME also impact treatment outcomes. SMARCA4-Mut tumors are coregulated by multiple key factors, and Napsin A may affect only one.
Conclusions
SMARCA4-Mut is an adverse prognostic feature in NSCLC patients. Napsin A expression in SMARCA4-Mut patients is associated with prolonged OS. However, we are aware of several limitations of our study, the most important of which are its retrospective type, small patient sample, and treatment heterogeneity. Thus, the clinical relevance of SMARCA4-Mut in NSCLC warrants further investigation.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-381/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-381/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-381/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-381/coif). J.J. serves as an unpaid editorial board member of Translational Lung Cancer Research from October 2023 to September 2025. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved granted by the Institutional Ethics Committee of the 900th Hospital of the Joint Logistic Support Force, People’s Liberation Army of China (No. 2021-026).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Carbone DP, Gandara DR, Antonia SJ, et al. Non-Small-Cell Lung Cancer: Role of the Immune System and Potential for Immunotherapy. J Thorac Oncol 2015;10:974-84. [Crossref] [PubMed]
- Relli V, Trerotola M, Guerra E, et al. Abandoning the Notion of Non-Small Cell Lung Cancer. Trends Mol Med 2019;25:585-94. [Crossref] [PubMed]
- Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J Thorac Oncol 2022;17:362-87. [Crossref] [PubMed]
- Mardinian K, Adashek JJ, Botta GP, et al. SMARCA4: Implications of an Altered Chromatin-Remodeling Gene for Cancer Development and Therapy. Mol Cancer Ther 2021;20:2341-51. [Crossref] [PubMed]
- Schoenfeld AJ, Bandlamudi C, Lavery JA, et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin Cancer Res 2020;26:5701-8. [Crossref] [PubMed]
- Alessi JV, Ricciuti B, Spurr LF, et al. SMARCA4 and Other SWItch/Sucrose NonFermentable Family Genomic Alterations in NSCLC: Clinicopathologic Characteristics and Outcomes to Immune Checkpoint Inhibition. J Thorac Oncol 2021;16:1176-87. [Crossref] [PubMed]
- Chatzopoulos K, Boland JM. Update on genetically defined lung neoplasms: NUT carcinoma and thoracic SMARCA4-deficient undifferentiated tumors. Virchows Arch 2021;478:21-30. [Crossref] [PubMed]
- Nambirajan A, Jain D. Recent updates in thoracic SMARCA4-deficient undifferentiated tumor. Semin Diagn Pathol 2021;38:83-9. [Crossref] [PubMed]
- Naito T, Udagawa H, Umemura S, et al. Non-small cell lung cancer with loss of expression of the SWI/SNF complex is associated with aggressive clinicopathological features, PD-L1-positive status, and high tumor mutation burden. Lung Cancer 2019;138:35-42. [Crossref] [PubMed]
- Tian Y, Xu L, Li X, et al. SMARCA4: Current status and future perspectives in non-small-cell lung cancer. Cancer Lett 2023;554:216022. [Crossref] [PubMed]
- Takada K, Sugita S, Murase K, et al. Exceptionally rapid response to pembrolizumab in a SMARCA4-deficient thoracic sarcoma overexpressing PD-L1: A case report. Thorac Cancer 2019;10:2312-5. Erratum in: Thorac Cancer 2020;11:3645. [Crossref] [PubMed]
- Li HR, Gao J, Jin C, et al. Downregulation of SETBP1 promoted non-small cell lung cancer progression by inducing cellular EMT and disordered immune status. Am J Transl Res 2020;12:447-62. [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Jiao S, Zhang X, Wang D, et al. Genetic Alteration and Their Significance on Clinical Events in Small Cell Lung Cancer. Cancer Manag Res 2022;14:1493-505. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Walter DM, Venancio OS, Buza EL, et al. Systematic In Vivo Inactivation of Chromatin-Regulating Enzymes Identifies Setd2 as a Potent Tumor Suppressor in Lung Adenocarcinoma. Cancer Res 2017;77:1719-29. [Crossref] [PubMed]
- Lissanu Deribe Y, Sun Y, Terranova C, et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nat Med 2018;24:1047-57. [Crossref] [PubMed]
- Concepcion CP, Ma S, LaFave LM, et al. Smarca4 Inactivation Promotes Lineage-Specific Transformation and Early Metastatic Features in the Lung. Cancer Discov 2022;12:562-85. [Crossref] [PubMed]
- Roh H, Lee SY, Lee J, et al. Use of thyroid transcription factor 1 and napsin A to predict local failure and survival after Gamma Knife radiosurgery in patients with brain metastases from lung adenocarcinoma. J Neurosurg 2023;138:663-73. [Crossref] [PubMed]
- Li F, Wang S, Wang Y, et al. Multi-omics analysis unravels the underlying mechanisms of poor prognosis and differential therapeutic responses of solid predominant lung adenocarcinoma. Front Immunol 2023;14:1101649. [Crossref] [PubMed]
- Xue Y, Morris JL, Yang K, et al. SMARCA4/2 loss inhibits chemotherapy-induced apoptosis by restricting IP3R3-mediated Ca(2+) flux to mitochondria. Nat Commun 2021;12:5404. [Crossref] [PubMed]
- Morrison AJ. Chromatin-remodeling links metabolic signaling to gene expression. Mol Metab 2020;38:100973. [Crossref] [PubMed]
- Zhu X, Fu Z, Chen SY, et al. Alanine supplementation exploits glutamine dependency induced by SMARCA4/2-loss. Nat Commun 2023;14:2894. [Crossref] [PubMed]


