
IL-1 receptor-associated kinase 3 (IRAK3) in lung adenocarcinoma predicts prognosis and immunotherapy resistance: involvement of multiple inflammation-related pathways
Highlight box
Key findings
• Reduced IL-1 receptor-associated kinase 3 (IRAK3) protein level was observed to correlate with unfavorable prognosis for lung adenocarcinoma. Elevated IRAK3 exacerbated cytotoxic T lymphocyte dysfunction and predicted immunotherapy resistance through multiple inflammation-related pathways.
What is known and what is new?
• In previous studies, IRAK3 has been suggested as a suppressor of innate immune signaling and IRAK3 exhibits controversial roles in tumor progression, patient prognosis, immune evasion and therapy efficacy in diverse cancer types.
• In this study, we examined IRAK3 protein level and its prognostic value in lung adenocarcinoma (LUAD) by our National Cancer Center cohort. Moreover, elevated IRAK3 could promote immunotherapy resistance and increase response to chemotherapeutic and molecular targeted drugs.
What is the implication, and what should change now?
• Considering the clinical and immune relevance of IRAK3 in LUAD, we have the opportunity to select more effective treatments based on IRAK3 level in LUAD tissues. Further researches are warranted to make IRAK3 a plausible therapeutic target for LUAD patients.
Introduction
Lung cancer, with about 18.0% of all cancer mortality, stands as the primary contributor to cancer-related death on a global scale and remains the major global health challenge (1). Lung adenocarcinoma (LUAD) is the predominant histological type of lung cancer, representing over 40% of all diagnosed lung cancer cases (2). Currently, the prevailing and crucial method for predicting prognoses in LUAD patients is the accurate staging utilizing the tumor-node-metastasis (TNM) staging system (3). Nevertheless, tumor heterogeneity compromises the predictive accuracy of TNM staging, which suggests that novel molecular prognostic markers should be identified to determine risk stratification and prognoses (4-6). Besides promising molecular markers in tumor cells, tumor microenvironment (TME), comprised of tumor-infiltrating immune cells (TICs) and stromal cells, has appealed to more scientists. Recent researches have indicated that specific categories of TICs are linked to the prognosis of LUAD (7,8).
Although significant progresses have been made in the comprehensive treatment of LUAD, the sensitivity to chemotherapeutic and molecular targeted agents varies extensively among patients (9-11). Immunotherapeutic strategies, such as programmed death ligand-1 (PD-L1), programmed death-1 (PD-1) and cytotoxic T lymphocyte associated antigen 4 (CTLA4) inhibitors, are considered as vital parts of conventional treatment and showed promising efficacy in LUAD (12,13). Nevertheless, only a minor proportion of LUAD patients experience a sustained response due to tumor cells heterogeneity and variable immune phenotypes (14,15). In addition, tumor-infiltrating T lymphocytes (16-19) and other immune cells including immune suppressive macrophages (20), plasma cells (21,22) and myeloid-derived suppressor cells (MDSCs) (23), are reported to impact the clinical outcome and effectiveness of both chemotherapy and immunotherapy (24). Hence, a better understanding of interplays between immune system and tumor cells, and further elucidation of immune phenotypes are urgently needed for predicting clinical therapy responses and identifying novel therapeutic targets in LUAD.
IL-1 receptor-associated kinase 3 (IRAK3) belongs to the family of IL-1 receptor-associated kinases, a group that has pivotal functions in innate immune signaling. As opposed to other members of the IRAK family, IRAK3 is believed to be catalytically inactive and negatively regulate the TLR or IL-1R signaling pathways (25). IRAK3 stabilizes the Myddosome complex and prevents the separation of IRAK1/IRAK4 from the TLR receptor, hindering the assembly of IRAK1/TRAF6 complexes and thus suppressing downstream signaling cascades and impeding the activation of various pathways including AKT, NF-κB and MAPK/ERK (26). IRAK3 predominantly resides in myeloid cells such as monocytes and macrophages (27). Most published researches are focusing on IRAK3 expression in myeloid cells and it is widely accepted that cancer-derived cytokines could induce IRAK3 expression in human myeloid cells causing monocyte/macrophage deactivation and tumor immune evasion (28-30). In terms of the intrinsic roles of IRAK3 in cancer cells, only few studies were conducted and controversial perspectives have been proposed due to differential levels of IRKA3 in diverse cancers. In colorectal cancer, tumor cell-intrinsic IRAK3 promotes tumor progression by regulating antimicrobial response and STAT3 stability (31,32). In contrast, IRAK3 is deficient or decreased in more cancer types such as melanoma, glioma and hepatocellular carcinoma (27,33). Further results suggest that restoration of IRAK3 in IRAK3-deficient cancers inhibits tumor growth and induces cell death (34). With clinical data from multiple cohorts, expression level and epigenetic alteration of IRAK3 are found to be predictive of survival, diagnosis or treatment response in patients with various cancers (34-38). These results underscore the potentials of IRAK3 as a prognostic predictor and a novel immunotherapeutic target. However, its roles in LUAD progression and survival of LUAD patients are unclear. Less is known about how IRAK3 impacts the tumor immune infiltration and regulates responses to immune therapy among patients.
This research extensively examined the genetic expression and epigenetic alteration of IRAK3 in multiple cancer types. We analyzed IRAK3 protein level and its relevance with prognosis in LUAD utilizing both online data resources and our National Cancer Center (NCC) cohort. The purpose of this research was to determine whether reduced IRAK3 occurs in LUAD and whether mRNA or protein levels of IRAK3 have any relationship with clinical outcomes of LUAD patients. Based on prior researches about intrinsic roles of IRAK3 in other cancer types, the prespecified hypothesis tested was that reduced protein expression levels in the tumor are related with shorter overall survival (OS) in LUAD. Moreover, we explored the correlation between IRAK3 expression and TICs in TME by six advanced algorithms. The potentials of IRAK3 to predict the immunotherapy and chemotherapy responses in LUAD patients were also investigated. These studies are significant as they shed light on the critical roles of IRAK3 in the tumor-immune interactions of LUAD and offer the opportunity to select more effective treatments based on IRAK3 levels in tumor tissues. We present this article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-391/rc).
Methods
Database analysis and data collection
In the Oncomine database (https://www.oncomine.org, retrieved on September 21st, 2021), we evaluated mRNA expression levels of the IRAK3 gene across diverse cancer types and their corresponding normal tissues (39). The threshold was set based on the values provided below: gene ranking of the top 10% genes, P value of 0.0001 and fold change of 2. TIMER database (https://cistrome.shinyapps.io/timer/) provides a comprehensive platform for the systematical analysis of immune infiltration in a variety of cancer types (40). We visualized IRAK3 expression sourced from different types of cancer and their corresponding normal tissues in The Cancer Genome Atlas (TCGA) via Diff Exp module. In addition, mRNA expression, DNA methylation data, survival outcomes and clinical information of all available human cancer types within TCGA database was obtained from the UCSC Xena website (https://xenabrowser.net/datapages/) (41). TCGA data was utilized to examine differences in the methylation status of IRAK3 among various cancer types. We extracted all CpG sites methylation status of IRAK3 to evaluate the correlation between IRAK3 expression and its methylation level.
Patient samples
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study design and protocol were approved by the Research Ethics Committee of the NCC/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC2020C-506, approved on 17 November 2020) and individual consent for this retrospective analysis was waived. We retrospectively obtained formalin-fixed paraffin-embedded (FFPE) cancer tissue specimens of 250 LUAD patients, as well as their matched non-tumor tissues, which were retrieved from the biobank of the NCC (China). Though cost and practical issues restricted the sample size of our study, the analysis of tissue samples from over 200 patients had 90% power to detect an absolute difference of 10% in OS associated with IRAK3 level. We recruited participants based on inclusion criteria including primary diagnosis of LUAD between 2006 and 2017, no previous diagnosis of carcinoma, no metastatic disease at diagnosis, tumor stage I/II/III and surgical excision without any preoperative treatment. Patients who experienced neoadjuvant treatment and inoperable stage IV patients were excluded. Patients were treated with surgery by either minimally invasive sub-lobectomy (109 cases), lobectomy (133 cases) or pneumonectomy (8 cases) with lymph nodes resection or sampling. Original specimens were resected surgically from enrolled LUAD patients and instantly fixed in 10% buffered formalin for 24 h, dehydrated in 70% ethanol (EtOH) and paraffin embedded. FFPE sections were cut by a cryostat (Leica Microsystems, Milton Keynes, UK) and stored at room temperature for further analysis. Following adjuvant treatment was based on treatment guidelines which varied over time. The clinical information about smoking history, age and gender, tumor size, tumor node metastasis stage and OS was retrieved from medical records and follow-up surveys.
Immunohistochemistry staining and scoring
Preparations of paraffin-embedded sections from LUAD tissues and adjacent non-tumor tissues were completed for immunohistochemical staining. In brief, the sections were subjected to microwaving for antigen retrieval in antigen retrieval buffer with pH 6.0. Slides were incubated with anti-IRAK3 (HPA043097, Sigma-Aldrich, Darmstadt, Germany) antibody at 4 ℃ overnight, followed by incubation with the secondary antibody. Two pathologists (W.R. and Zhao Li) independently scored the slides, considering both the staining intensity and the percentage of cells showing positive staining. Their scoring process was blinded to clinical data and the agreement between them was over 90%. Discordant cases were reviewed by a senior experienced pathologist and were reassigned on consensus of opinion. The staining intensity was categorized into four levels as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). The percentage of staining was assessed using the following scale: 0 (negative), 1 (<10%), 2 (10–50%), 3 (51–80%), or 4 (>80%). The immunohistochemistry (IHC) index, a result of merging the staining intensity and proportion scores, was utilized to determine expression levels. In the absence of the clinically established cutoff point for IRAK3, patients were categorized into high (IHC index ≥6) and low (IHC index <6) groups based on the median.
Survival prognosis analysis
All patients from the NCC LUAD cohort were examined routinely every 6 months during the first 5 years of follow-up and once a year thereafter. The estimated median follow-up time calculated by the reverse Kaplan-Meier method was 8.0 years. The primary endpoint was death of a patient. OS was defined as time from pulmonary operation to death of all causes. Progression-free survival (PFS) was defined as time from operation to the first progression including tumor recurrence, contralateral tumor, secondary tumor or death. Using survival data from the NCC LUAD cohort, along with IRAK3 expression level, we conducted survival analysis employing the Kaplan-Meier method and determined statistical significance with the log-rank test. We sought to determine independent prognostic factors by utilizing a Cox proportional hazards model. Hazard ratios (HRs) were calculated as the ratios of the hazard in the high-IRAK3 group to the hazard in the low-IRAK3 group, serving as a measure to evaluate the influence of IRAK3 on patient survival. The other considered variables for Cox survival analyses were age, gender, tobacco use, tumor size, histology grade and TNM stage. Simultaneous adjustments for these candidate variables were performed. Meanwhile, survival curves depicting OS in LUAD patients from our cohort, with or without lymphatic metastasis, were plotted respectively. The Kaplan-Meier plotter database (https://kmplot.com/analysis/) included 2,852 NSCLC specimens with an average follow-up of 49.7 months (42). The correlations between IRAK3 expression and survivals in lung cancers, lung squamous cell carcinomas, LUADs, LUAD with or without lymphatic metastasis were thoroughly investigated using Kaplan-Meier plotter. Corresponding survival curves, illustrating both OS and PFS, were graphically represented.
IRAK3 coexpression network and functional enrichment analysis
The LinkedOmics database (https://www.linkedomics.org/login.php) includes clinical information and multi-omics data from 10 Clinical Proteomics Tumor Analysis Consortium (CPTAC) cancer cohorts and all 32 TCGA cancer types. LinkedOmics serves as an exclusive and accessible platform enabling clinicians and biologists to combine and analyze multi-omics data within and across tumor types (43). We identified differentially expressed genes associated with IRAK3 using the LinkFinder module in the TCGA LUAD cohort. Functional enrichment analyses based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) functional annotations were performed in the LinkInterpreter module. Besides, we applied the ‘clusterProfiler’ package to complete the GSEA and identify the pathways that are linked to IRAK3-related genes. Corresponding line plots were generated by the ‘enrichplot’ package in R.
IRAK3 and tumor immune microenvironment
The TIMER2.0 (http://timer.comp-genomics.org/timer/, accessed on 20 May 2023) database provides an extensive platform for systematically analyzing immune infiltrates in various cancer types (44). Six advanced algorithms including CIBERSORT, XCELL, EPIC, QUANTISEQ, MCPCOUNTER and TIMER are all applied to rigorously evaluate the levels of immune cell infiltration within tumors based on TCGA data. We explored the relationship between IRAK3 and six types of immune cell infiltration levels by Spearman’s correlation test. Besides, we also calculated the exact proportion of diverse immune cell types in every LUAD specimen by the ‘CIBERSORT’ package (45). In order to assess tumor purity and verify the extent of infiltration by stromal and immune cells, we quantified ESTIMATE, immune and stromal scores of each LUAD specimen by the ‘ESTIMATE’ R package (46). Spearman correlation analyses were conducted to calculate correlation coefficients between IRAK3 and ESTIMATE scores, immune scores and stromal scores.
IRAK3 expression at the single-cell level
The Tumor Immune Single-cell Hub 2 (TISCH2) (http://tisch.comp-genomics.org/) serves as an extensive and up-to-date repository of single-cell RNA-seq (scRNA-seq) data which enables the single-cell transcriptome exploration and interactive visualization of TME across multiple cancer types (47). TISCH2 has integrated 190 scRNA-seq datasets from tumors, encompassing over 6 million cells among various cancer types. At the single-cell level, IRAK3 expressions in both LUAD_GSE131907 dataset [Gene Expression Omnibus (GEO) accession: GSE131907] (48) and LUAD_GSE149655 dataset (GEO accession: GSE149655) (49) were visualized via dataset module. Heatmaps of IRAK3 expression from diverse cellular types of all NSCLC datasets were achieved through gene module. Besides, correlation analyses were utilized to calculate correlation coefficients between IRAK3 and myeloid cell lineage-specific genes including CD45, CD11b and CD11c.
IRAK3 and immunotherapy response
The Tumor Immune Dysfunction and Exclusion (TIDE) database (http://tide.dfci.harvard.edu/) is an interactive and comprehensive repository and its algorithm behind the computational method is designed to predict immunotherapy responses and to evaluate immune escape potentials of different tumors according to gene expression profiles (50,51). The TCGA-LUAD cohort was utilized to examine the effect of IRAK3 on the responses to immunotherapy in tumor cases. TIDE scores, T-cell dysfunction scores and exclusion scores of each patient were acquired by TIDE algorithm. Among them, TIDE scores were used for immunotherapy response prediction and tumor immune evasion evaluation. T-cell dysfunction scores were utilized to examine how the levels of cytotoxic T lymphocytes (CTLs) correlate with overall patient survival across various IRAK3 levels. Inversely correlated with dysfunction, T-cell exclusion scores were derived from gene signatures associated with immunosuppressive cells, which predict immune escape. Additionally, microsatellite instability (MSI) and tumor mutation burden (TMB) are well recognized as the indicators predicting the efficacy of immunotherapy in multiple cancers. We obtained simple nucleoside variation data from TCGA-LUAD cohort for the purpose of calculating the TMBs. The MSI of TCGA-LUAD cohort was also retrieved from TIDE database. Spearman correlation analyses were performed to evaluate the relationships between IRAK3 and immune checkpoint markers including BTLA, PDCD1, PDCD1LG2, CD274, HAVCR2, CTLA4, TIGIT and LAG3.
IRAK3 and drug response
The Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/) stands as the comprehensive and accessible repository providing potential biomarkers of drug response and details about multiple drug sensitivity in tumor cells (52). Both GDSC1 and GDSC2 datasets were utilized to evaluate how each patient responded various chemotherapeutic and molecular targeted agents (53,54). The half-maximal inhibitory concentration (IC50) values, representing the concentration at which inhibition is 50%, were calculated by the ‘oncoPredict’ package in the R programming environment (55).
Statistical analysis
We evaluated the statistics utilizing SPSS statistics 22 (SPSS, Chicago, IL, USA), GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA) or R software (version 4.3.1). Through the Chi-squared test, we assessed the connections between clinicopathological features and IRAK3 expression. Patient survival was analyzed with the Kaplan-Meier method and evaluated for statistical significance using the log-rank test. Besides, survival data was fitted with Cox proportional hazards models for the purpose of identifying independent prognostic factors. Both Pearson and Spearman methods were used for correlation analysis. The statistical significance test between different groups was completed by two-tailed Student’s t-tests, and the outcomes were reported as mean ± standard deviation (SD). Statistical significance was defined when P<0.05.
Results
IRAK3 is differentially expressed and frequently methylated in multiple human cancer types
To assess variations in IRAK3 expression between tumors and normal tissues, we thoroughly went through the Oncomine database and examined IRAK3 transcription levels across different types of cancer and their corresponding non-tumor tissues. This analysis indicated elevated IRAK3 expression only in a few datasets including colorectal, esophageal, kidney, pancreatic cancers, brain and central nervous system (CNS) cancer and lymphoma. Conversely, diminished IRAK3 expression was observed in more cancer types when compared to the normal tissues, such as lung, breast, bladder, gastric, ovarian, prostate cancers, leukemia, melanoma and sarcoma (Figure 1A).
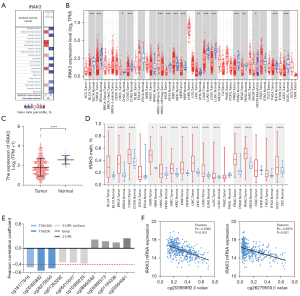
We utilized the TCGA data to conduct additional assessments of IRAK3 expression across various human cancers. After RNA-seq data extraction from TCGA, differential expressions of IRAK3 in multiple cancers and their adjacent normal tissues were illustrated in Figure 1B. Specifically, in the TCGA-LUAD cohort the mRNA expression of IRAK3 was found to be reduced in tumor tissues in contrast to their levels in adjacent normal tissues (Figure 1C). Additionally, we used methylation data from TCGA to predict whether the expression of IRAK3 is regulated by promoter region methylation. Methylation data of IRAK3 in human cancers were shown in Figure 1D. IRAK3 methylation levels in most human cancers are significantly elevated comparing to those of corresponding normal tissues. The increased methylation level showed a reverse trend to mRNA expression in multiple human cancer types. The IRAK3 expression in LUAD was inversely related to cg20395892 and cg26279550 site methylation, which are located in the promoter region (P<0.001, Figure 1E,1F). These findings suggest that IRAK3 expression regulation is influenced by promoter region methylation.
IRAK3 is significantly down-regulated in LUAD tissue and decreased IRAK3 level is associated with unfavorable prognosis
Tumor samples and their matched non-tumor tissue samples from 250 LUAD patients were examined for IRAK3 assays by immunohistochemistry staining, and evaluable assay results were generated for 222 patients (89%). Among the 28 dropouts, 16 (57%) had inadequate tumor cells, 7 (25%) had poor tumor preservation, 4 (14%) had insufficient control staining, and 1 (4%) had excessive necrosis. Then we evaluate the positive IRAK3 protein expression in 222 LUAD specimens and found a notable reduction of IRAK3 level within tumor tissues relative to matched normal tissues (Figure 2A,2B). The IHC staining results also suggest that the predominant localization of IRAK3 is within the cytoplasm, observed in both tumor and normal tissues (Figure 2B).
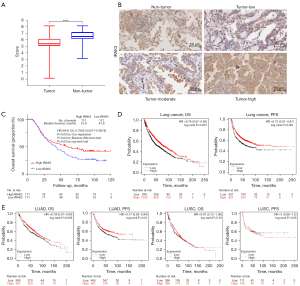
Moreover, the IRAK3 prognostic potential was examined in our LUAD cohort. The IRAK3 level exhibited a marked association with patient survival. According to both univariate Cox regression analysis (P=0.03, Table 1) and Kaplan-Meier analysis (P=0.03, log-rank test, Figure 2C), patients with low IRAK3 level experienced poorer outcomes. Independent prognostic predictors for patients with LUAD analyzed by the multivariate Cox proportional hazards regression were identified, including older age (HR =1.934, P<0.001), larger tumor size (HR =1.914, P<0.001), higher TNM stage (HR =1.614, P=0.006) and decreased IRAK3 protein level (HR =0.710, P=0.04) (Table 1). We further checked covariates for possible time-varying effects in the Cox regression model and no significant deviation from the proportional-hazard assumption could be found.
Table 1
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age (≥60 vs. <60 years) | 1.849 (1.330–2.571) | <0.001 | 1.934 (1.387–2.696) | <0.001 | |
| Gender (male vs. female) | 1.276 (0.916–1.778) | 0.15 | |||
| Tobacco use (yes vs. no) | 1.277 (0.924–1.763) | 0.14 | |||
| Tumor size (>4 vs. ≤4 cm) | 2.239 (1.615–3.104) | <0.001 | 1.914 (1.361–2.690) | <0.001 | |
| Histology grade (3 vs. 1/2) | 1.474 (1.059–2.052) | 0.02 | 1.223 (0.858–1.744) | 0.27 | |
| TNM stage (III vs. I/II) | 1.879 (1.358–2.599) | <0.001 | 1.614 (1.148–2.268) | 0.006 | |
| IRAK3 (high vs. low) | 0.700 (0.505–0.969) | 0.03 | 0.710 (0.512–0.984) | 0.04 | |
LUAD, lung adenocarcinoma; HR, hazard ratio; CI, confidence interval; TNM stage, tumor-node-metastasis stage.
In Kaplan-Meier plotter database, we further examined the relationship between IRAK3 expression and lung cancer prognosis. As illustrated in Figure 2D, the better prognosis in lung cancer [OS HR =0.76, 95% confidence interval (CI): 0.67 to 0.85, P<0.001; PFS HR =0.73, 95% CI: 0.61 to 0.87, P<0.001] was associated with higher IRAK3 expression. After more in-depth evaluation of IRAK3 prognostic potentials in pathological subtypes of lung cancers, our analysis revealed a notable association between IRAK3 expression and patient survival only in LUAD (Figure 2E). The results above confirmed the prognostic significance of IRAK3 in LUAD, highlighting that diminished IRAK3 level leads to poor prognosis.
IRAK3 expression level impacts the prognosis of LUAD patients with lymphatic metastasis
To gain deeper insights into the mechanisms underlying IRAK3’s role as a prognostic indicator in LUAD, we explored the correlations between IRAK3 levels and clinicopathological features of our LUAD cohort (Table 2). Results of the chi-square test indicated a notable association between IRAK3 levels and histology grade (P=0.03), TNM stage (P=0.041) and N stage (P=0.03). Interestingly, decreased IRAK3 was related with worse OS in stage N1 and N2 patients (OS HR =0.5519, P=0.02, log-rank test) but was not correlated with OS of N0 patients (OS HR =1.035, P=0.89, log-rank test) (Figure 3A,3B). In this context, the N category indicates lymph nodes involvement. Specifically, N0 denotes the absence of regional lymph node metastasis, while N1, N2 and N3 signify lymph node metastases of different sites (56).
Table 2
| Features | Number of cases | IRAK3 expression | P | |
|---|---|---|---|---|
| High | Low | |||
| Age, years | 0.42 | |||
| ≥60 | 118 | 62 | 56 | |
| <60 | 104 | 49 | 55 | |
| Gender | 0.34 | |||
| Male | 131 | 62 | 69 | |
| Female | 91 | 49 | 42 | |
| Tobacco use | 0.08 | |||
| Yes | 99 | 43 | 56 | |
| No | 123 | 68 | 55 | |
| Tumor size | 0.69 | |||
| >4 cm | 101 | 49 | 52 | |
| ≤4 cm | 121 | 62 | 59 | |
| Histology grade | 0.03 | |||
| G1 + G2 | 98 | 41 | 57 | |
| G3 | 124 | 70 | 54 | |
| TNM stage | 0.041 | |||
| I+II | 124 | 69 | 55 | |
| III | 98 | 41 | 57 | |
| T stage | 0.16 | |||
| 1+2 | 169 | 89 | 80 | |
| 3+4 | 53 | 22 | 31 | |
| N stage | 0.03 | |||
| 0 | 90 | 53 | 37 | |
| 1+2+3 | 132 | 58 | 74 | |
LUAD, lung adenocarcinoma; TNM stage, tumor-node-metastasis stage; T stage, tumor stage; N stage, node stage.
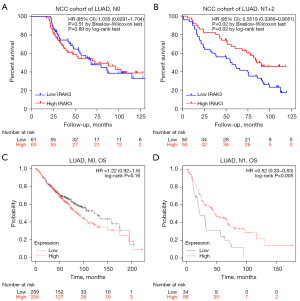
In the Kaplan-Meier plotter database, we further examined the relevance between IRAK3 expression and clinical outcomes in LUAD patients with various clinicopathological factors. Low expression of IRAK3 was found to correlate with unfavorable OS and PFS in both genders (P<0.05, Table S1). Moreover, IRAK3 expression level was verified to be markedly correlated with node-positive patient survival in the Kaplan-Meier plotter database. To be specific, IRAK3 expression levels were found to be irrelevant for N0 patients (OS HR =1.22, P=0.16, log-rank test, Figure 3C), while patients at stage N1 with reduced IRAK3 expression exhibited unfavorable outcomes, as indicated by Kaplan-Meier analysis (OS HR =0.52, P=0.005, log-rank test, Figure 3D). The findings imply that both mRNA expression and protein levels of IRAK3 has the potential to influence the prognosis of LUAD patients with lymph node metastasis.
IRAK3 coexpression network and IRAK3-related signaling pathways are identified in LUAD
In the LinkedOmics web portal, we utilized the LinkFinder module to illustrate the coexpression network of IRAK3 in TCGA-LUAD for the purpose of exploring its biological function in LUAD. As depicted in Figure 4A, 6,320 genes (highlighted in red dots) exhibited a positive correlation with IRAK3. Reversely, a total of 4,227 genes (highlighted in green dots) displayed a negative correlation with IRAK3. Among them, 50 most positively and 50 most negatively correlated genes generated heatmaps respectively (Figure 4B,4C). Almost all, specifically 48 out of the leading 50 genes positively correlated with IRAK3, demonstrated favorable HR values, which suggested great potential of becoming protective markers in LUAD. In contrast, the majority of the top 50 genes displaying negative correlation, were strongly indicative of being high-risk markers with unfavorable HR values (Figure 4D). The results above show that IRAK3 coexpression network could potentially influence patients’ outcome and prognosis in LUAD.
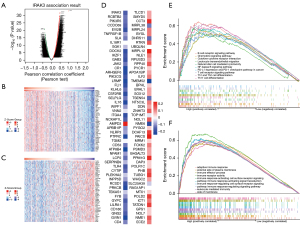
To pinpoint signaling pathways altered by IRAK3, we employed GSEA to detect the pathways activated in LUAD. The KEGG pathway enrichment analysis demonstrated that coexpressed genes associated with IRAK3 were mainly engaged in B cell receptor signaling pathway, chemokine signaling pathway, leukocyte transendothelial migration, nuclear factor kappa B (NF-κB) signaling pathway, cytokine-cytokine receptor interaction, PD-L1 expression and PD-1 checkpoint pathway in cancer, natural killer cell mediated cytotoxicity, T cell receptor signaling pathway, Th1 and Th2 cell differentiation and Th17 cell differentiation (Figure 4E). The outcomes of the GO term enrichment analysis revealed that coexpressed genes were mostly enriched in adaptive immune response, external side of plasma membrane, immune effector process, immune receptor activity, immune response-activating cell surface receptor signaling pathway, immune response-regulating signaling pathway, immune response-activating signal transduction, immune response-regulating cell surface receptor signaling pathway, leukocyte mediated immunity, etc. (Figure 4F). All the GSEA results suggest that IRAK3 and its coexpressed genes have potential roles in triggering and regulating immune responses in LUAD.
Correlation analysis between IRAK3 and immune infiltration levels in LUAD
In diverse cancers, the tumor-infiltrating lymphocyte level has been claimed to independently predict sentinel lymph node status and clinical outcomes (57,58). Hence, we examined the potential correlation between IRAK3 and the extent of immune infiltration in LUAD. We utilized CIBERSORT analysis to examine disparities in immune infiltration between IRAK3 low and high tumors. LUAD patients exhibiting high IRAK3 expression demonstrated markedly elevated levels of immune cell infiltration compared to those with low IRAK3 expression (Figure 5A). The Spearman correlation was utilized to assess the connection between IRAK3 expression and the observed immune infiltration levels, and IRAK3 exhibited a direct positive correlation with the abundance of macrophages, CD4+ T cells, CD8+ T cells, neutrophils, monocytes, etc. (P<0.001, Figure 5B-5D). Besides, we adopted a series of algorithms including EPIC, MCPCOUNTER, QUANTISEQ, XCELL and TIMER to evaluate immune infiltration levels in LUAD. The outcomes from these algorithms and the TISIDB database demonstrated positive correlations between IRAK3 and several immune cell types, particularly macrophages, monocytes, CD4+ T cells, myeloid dendritic cells and B cells, which corresponded with the result of CIBERSORT analysis (Figure S1A-S1C). Furthermore, our findings indicated a notable elevation in the ESTIMATE, immune and stromal scores in LUAD caused by IRAK3 stimulation (Figure 5E). All the above results strongly indicate a close connection between IRAK3 and immune infiltration within the TME.
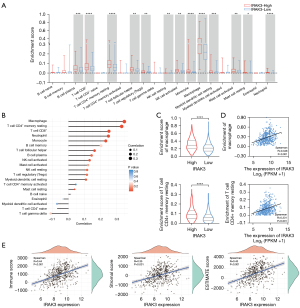
IRAK3 specific expression on monocyte/macrophage
For purpose of investigating how IRAK3 impacts tumor immune microenvironment, we extracted publicly available scRNA-seq data to examine IRAK3 expression levels across various immune and stromal cell types within the LUAD microenvironment. We discovered that IRAK3 was predominately expressed on monocyte/macrophage, endothelial cells and fibroblasts (Figure 6A-6C). Furthermore, IRAK3 expression exhibited positive correlations with immune cell markers including CD45 (R=0.442, P<0.001), CD11b (R=0.362, P<0.001) and CD11c (R=0.402, P<0.001) in selected LUAD datasets (Figure 6D). The robust correlations observed suggest that IRAK3 is predominantly expressed by myeloid cells, including both monocytes and macrophages.
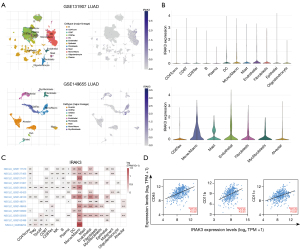
Correlation between IRAK3 expression and immunotherapy response
Immunotherapy for cancer utilizing immune checkpoint blockade (ICB) can activate immune system to detect and eradicate tumor cells, but ICB is effective in only about one-third of patients across various cancer types (59,60). Identification of sensitive and reliable ICB response biomarkers is crucial for individuals with tumors. We utilized the TIDE algorithm to examine predictive responses to immunotherapy among TCGA-LUAD patients. Our findings revealed a notable decrease in IRAK3 expression among LUAD patients who exhibited a heightened responsiveness to immunotherapy, as opposed to those who demonstrated resistance to immunotherapy (Figure 7A). The TIDE scores showed a marked increase in the high-IRAK3 group when contrasted with the low-IRAK3 group (P<0.001) and we observed elevated TMB and MSI in the low-IRAK3 group (P<0.001 and P<0.001, respectively, Figure 7B,7C). This suggests that LUAD patients with elevated IRAK3 levels are more inclined to be unresponsive to ICB treatment in contrast to patients with lower IRAK3 expression. Additionally, the high-IRAK3 group exhibited higher T-cell dysfunction scores (P<0.001, Figure7C). These findings implied that IRAK3 expression was connected with the evasion of tumor immunity and resistance to immunotherapy. Targeting IRAK3 may weaken T-cell dysfunction and increase MSI and TMB, thereby improving the ICB response rate.
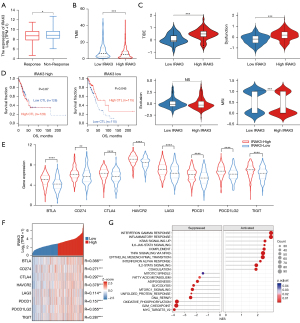
We delved deeper into understanding the mechanism by which IRAK3 promotes resistance to immunotherapy using the TIDE database (50). CTLs are major participants in the anti-tumor immune response, although their ability to control tumor growth may be compromised when they are in a dysfunctional state (61). In TCGA LUAD cohort, enhanced CTL levels correlated with improved patient survival, specifically when IRAK3 exhibited low expression. However, elevated IRAK3 expression attenuated or even reversed the positive impact of CTL (Figure 7D). These results suggested that IRAK3 exacerbates dysfunction in CTLs, contributing to the promotion of resistance to immunotherapy. All immune checkpoint markers were markedly elevated in high-IRAK3 patients, which implied a connection between IRAK3 expression and both immune escape and responses to immunotherapy (Figure 7E). The gene coexpression analysis also revealed positive correlations between IRAK3 and the expression of immune checkpoint markers, including PDCD1 (PD-1), CD274 (PD-L1), CTLA4, LAG3 and TIGIT (Figure 7F). Moreover, hallmark gene sets enrichment analysis indicated that IRAK3 significantly activated inflammatory response, interferon-gamma (IFN-γ) response and IL-6/JAK/STAT3 signaling pathways (Figure 7G, Figure S2A-S2C). Besides, the correlations of IRAK3 with essential components of the aforementioned pathways contributing to immunotherapy resistance were validated (Figure S2D).
Predicting the sensitivity of patients to antitumor drugs
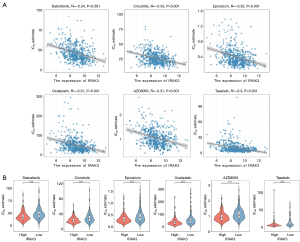
We computed IC50 values for various molecular targeted and chemotherapeutic agents commonly employed in treating LUAD through the ‘oncoPredict’ package. The calculated values were aimed at the treatment response prediction of patients with diverse levels of IRAK3 expression. The findings indicated that IRAK3 was notably inversely correlated with IC50 of dabrafenib (R=−0.34, P<0.001), crizotinib (R=−0.33, P<0.001), epirubicin (R=−0.32, P<0.001), oxaliplatin (R=−0.31, P<0.001), AZD8055 (R=−0.31, P<0.001) and taselisib (R=−0.30, P<0.001) (Figure 8A). The high-IRAK3 group exhibited significantly lower IC50 values for the anti-tumor drugs, indicating more favorable responses to the treatment (Figure 8B). These findings provided valuable insights for tailoring treatment strategies for patients with high IRAK3 expression.
Discussion
Despite being discovered more than 20 years ago, IRAK3 is generally perceived as a catalytically inactive pseudokinase, lacking crucial active site residues (62,63). Although IRAK3 has not been investigated thoroughly, IRAK3 is widely regarded as a suppressor of innate immune signaling and alterations of IRAK3 are linked to the onset and progression of tumor (25-27). Current studies have indicated that IRAK3 gene exhibits varied expression patterns in different cancer types and plays diverse roles in tumorigenesis, tumor progression, immune evasion, patients’ prognosis and therapy responsiveness (28,29,31,33-38). However, the roles of IRAK3 in LUAD and its underlying mechanisms remain inadequately elucidated.
Due to the discrepant distribution of IRAK3 in diverse cancers, we reported variations of IRAK3 expression and methylation levels in different types of cancers. Most cancer types are generally deficient in IRAK3 than paired normal tissues. Only a few cancer types including esophageal cancer, colorectal cancer, gastric cancer and pancreatic cancer reveal relatively high IRAK3 expression in tumor tissues, which are consistent with findings of former researches (27,31,36). In addition, we discovered reversed methylation level of IRAK3 in most cancer types. Inverse correlations between IRAK3 expression and methylation levels in LUAD provide insights into the regulation mechanisms at the transcriptional level. We proposed that DNA methylation of the promoter region located in the IRAK3 gene could be involved in regulating IRAK3 expression in LUAD.
With the LUAD cohort established in NCC/Cancer Hospital, we managed to use our data to verify IRAK3 protein levels in LUAD and evaluate its prognostic potential. Through IHC staining, IRAK3 was down-regulated in LUAD tissues, and elevated IRAK3 level exhibited a significant correlation with favorable OS among patients. It is worth noting that our finding contradicts a previously published study which claimed that IRAK3 expression in LUAD was correlated with unfavorable survival outcomes (28). Given that they used a multi-site cohort (GSE68465) of 439 LUAD patients (64), we applied Kaplan-Meier plotter database which included GSE68465 and other 16 independent cohorts to reexamine the IRAK3 prognostic potential (42). The survival analysis of integrated data demonstrated that lower IRAK3 expression led to worse prognosis, which is consistent with our finding about IRAK3 prognostic value. With variabilities in race, ethnicity, tumor staging, type of treatment, response to therapy, post-transcriptional regulation and even sample processing, the controversy of IRAK3 prognostic potential should be explainable. Meanwhile we cannot exclude the probability of multiple biases including publication bias, selection bias, information bias and confounding factors which occasionally occurred in retrospective cohort studies. Our finding distinguishes itself by addressing a broader spectrum of confounding variables compared with the prior study (Table 1). Besides age, gender, TNM stage and IRAK3 level, our study integrated tobacco use, tumor size and histology grade into the analysis. It turned out that age, tumor size, TNM stage and decreased IRAK3 level were factors significantly correlated with worse OS according to the multivariable analysis. Furthermore, it is interesting to find that both IRAK3 expression and its protein level were significantly associated with lymphatic metastasis and they had prognostic value only in patients with lymph node metastases by subgroup analysis. These results suggest that IRAK3 may function as a predictive marker for tumor metastasis, which offers insights in understanding the possible mechanisms pertinent to its role in prognosis prediction.
In the study, we explored IRAK3 gene coexpression network and IRAK3-related signaling pathways to find out molecular processes in which IRAK3 are involved. Aligning with the impact of IRAK3 on patient prognosis, 48 out of the leading 50 co-expressed genes showed lower risk of death. Survival maps of genes closely related to IRAK3 confirmed IRAK3’s prognostic value again. As mentioned previously, IRAK3 typically functions as a suppressor of NF-κB activation in innate immune signaling which results in a broad spectrum of proinflammatory cytokines and apoptosis-related factors (27,34,65,66). Besides that, all gene set enrichment analyses indicate that IRAK3 and its related genes are widely involved in multiple pathways linked to adaptive immune response. Specifically, T cell and B cell receptor signaling pathways, immune response-activating signal transduction pathways, immune response-regulating signaling pathways and even PD-L1 expression and PD-1 checkpoint pathway in cancer are all identified as IRAK3-related signaling pathways. Though experiment validation on these issues is pending, the new findings based on bioinformatic analysis enlightened our further exploration about IRAK3’s participations in tumor immune infiltration and immunotherapy response.
The development of tumors involves the participation of both innate and adaptive immune cells, whose interactions with cancer cells shape the TME, playing crucial roles in tumor progression, lymphatic metastasis, and evaluations of prognoses and treatment response (57,58,67-76). As mentioned in previous researches, IRAK3 expression is needed in above pathological process and clinical outcome. However, relationships between the IRAK3 expression and immune infiltrations in LUAD remain ambiguous. Our study showed that macrophages, CD4+ T cells and B cells were markedly positively correlated to IRAK3 expression. Macrophages and B cells have been proved to be linked to the prognosis of early-stage LUAD (7). Recent researches have indicated that macrophages and CD4+ T cells within the TME independently correlate with metastasis, recurrence and prognosis in stage I LUAD patients (77). Moreover, ESTIMATE, immune and stromal scores in patients with LUAD were promoted by IRAK3 level. Our results provided possible immune mechanisms by which IRAK3 influence the prognosis of LUAD, and bases for future in-depth studies of the immunological effects of IRAK3.
Being the most relevant immune cell type to IRAK3, monocyte/macrophage was found to express the maximum amount of IRAK3 in multiple LUAD datasets. Our finding verified that IRAK3 is chiefly expressed in monocyte and macrophage populations, which was in accordance with previous studies (28,29,78). Combined with above immune infiltration analysis, our results were consistent with a recent delicate study demonstrating that IRAK3 acted as an immune checkpoint specifically expressed in myeloid cells (78). In Irak3 knockout (KO) murine tumor models, Irak3 deficiency was proved to stimulate myeloid cells activation, leading to enhanced antigen-presentation capacity of these cells (79). Another in vivo mouse study has indicated that tumor-associated macrophages exhibit increased IRAK3 expression and defective cytokine production (28). We propose that IRAK3 is a crucial immune regulator in monocyte/macrophage and a cross-talk mediator between macrophages and cancer cells. Additional investigations are warranted to elucidate the underlying mechanism how macrophage was regulated by IRAK3.
While our research has shed light on the clinical and immune relevance of IRAK3 in LUAD, there is scarce evidence to demonstrate its predictive and therapeutic potentials in cancer immunotherapy and chemotherapy. During the past decade, cancer therapy has been revolutionized by immunotherapy, but the efficacy of ICB remains limited and acquired resistance to ICB occurs inevitably in most patients with cancer (14,59,60,80,81). In the TME, IFN-γ secreted by immune cells has been observed to upregulate PD-L1 on cancer cells, which induces T cell exhaustion and CTL dysfunction, hence leading to tumor immune evasion (82-84). Besides that, sustained IFN-γ signaling could drive PD-L1 independent adaptive resistance through STAT1-related transcriptomic and epigenomic regulations, resulting in enhanced expression of interferon stimulated genes (ISGs) and increased ligands for various inhibitory receptors on T cells (85). Tumor cell-induced inflammation drives an immune-suppressive environment in most cases. Tumor intrinsic NLRP3 inflammasomes could be activated by PD-1 blockade, ultimately leading to dampened antitumor immune response and adaptive resistance to immunotherapy (86). In most tumor patients with chronic inflammatory conditions, the IL-6/JAK/STAT3 signaling pathway tends to be excessively activated, which can inhibit anti-tumor immune response by inducing immunosuppression (87). Our GSEA confirmed that inflammatory response, IFN-γ response and IL-6/JAK/STAT3 signaling were all notably activated in the group with elevated levels of IRAK3 (Figure S2). The analysis of gene co-expression revealed strong correlations between IRAK3 and immune checkpoint markers including PD-1, PD-L1, CTLA4, LAG3 and TIGIT. Due to the above reasons, we concluded that IRAK3 exacerbated CTL dysfunction via various pathways to promote resistance of tumor immunotherapy, which corroborated the results anticipated by the TIDE database. Moreover, we used the latest ‘oncoPredict’ algorithm to evaluate antitumor response of molecular targeted and chemotherapeutic agents based on IRAK3 expression. It is interesting to find that high IRAK3 expression is related to better responses of most antitumor drugs, which is in contrast to the conclusion we get in the immunotherapy. IRAK3-regulated inflammation plays dual roles in immunotherapy and chemotherapy. The contradicted IRAK3-related mechanisms elicited by immunotherapy and chemotherapy are still elusive. Despite being controversial, our results provided personalized therapeutic guidance for the management of LUAD patients with varying IRAK3 expression levels.
Even though we have unveiled the critical roles of IRAK3 in tumor-immune interactions of LUAD, our research does have certain constraints and limitations. Firstly, our bioinformatics analyses relied heavily on data obtained from publicly available databases. We still need to conduct invitro biological experiments and recruit more clinical samples to confirm our results and promote clinical implementation. Secondly, the underlying mechanisms and related pathways of IRAK3 in cancer progression and therapy resistance were not experimentally investigated in present study. The ongoing further research will ultimately elucidate the specific molecular role of IRAK3 in the process of tumorigenesis and therapy resistance, which will contribute to accurate prognosis prediction and optimized cancer therapy.
Conclusions
In summary, our study presents the inaugural investigation demonstrating the down-regulation of IRAK3 in LUAD tumor tissues and its significant correlation with prognosis and immune infiltration. Further analysis suggests that IRAK3 exacerbates CTLs dysfunction and thus predicts immunotherapy resistance through multiple inflammation-related pathways activation. Patients with higher IRAK3 expression are predicted to be more responsive to molecular targeted and chemotherapeutic agents used in LUAD. Our findings therefore emphasize the potential dual roles of IRAK3 as a promising prognostic marker and a plausible therapeutic target for LUAD patients.
Acknowledgments
We are grateful to all individuals who took part in this research.
Funding: This research was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-391/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-391/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-391/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-391/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Research Ethics Committee of the NCC/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC2020C-506, approved on 17 November 2020) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Lim W, Ridge CA, Nicholson AG, et al. The 8(th) lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg 2018;8:709-18. [Crossref] [PubMed]
- Song Y, Kelava L, Kiss I. MiRNAs in Lung Adenocarcinoma: Role, Diagnosis, Prognosis, and Therapy. Int J Mol Sci 2023;24:13302. [Crossref] [PubMed]
- Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 2013;368:1101-10. [Crossref] [PubMed]
- Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol 2009;4:22-9. [Crossref] [PubMed]
- Liu X, Wu S, Yang Y, et al. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed Pharmacother 2017;95:55-61. [Crossref] [PubMed]
- Li HX, Wang SQ, Lian ZX, et al. Relationship between Tumor Infiltrating Immune Cells and Tumor Metastasis and Its Prognostic Value in Cancer. Cells 2022;12:64. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Wang DC, Wang W, Zhu B, et al. Lung Cancer Heterogeneity and New Strategies for Drug Therapy. Annu Rev Pharmacol Toxicol 2018;58:531-46. [Crossref] [PubMed]
- Nair NU, Greninger P, Zhang X, et al. A landscape of response to drug combinations in non-small cell lung cancer. Nat Commun 2023;14:3830. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Yang CY, Yang JC, Yang PC. Precision Management of Advanced Non-Small Cell Lung Cancer. Annu Rev Med 2020;71:117-36. [Crossref] [PubMed]
- Hegde PS, Chen DS. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020;52:17-35. [Crossref] [PubMed]
- Liu L, Bai H, Wang C, et al. Efficacy and Safety of First-Line Immunotherapy Combinations for Advanced NSCLC: A Systematic Review and Network Meta-Analysis. J Thorac Oncol 2021;16:1099-117. [Crossref] [PubMed]
- Bremnes RM, Busund LT, Kilvær TL, et al. The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:789-800. [Crossref] [PubMed]
- Boulle G, Velut Y, Mansuet-Lupo A, et al. Chemoradiotherapy efficacy is predicted by intra-tumour CD8+/FoxP3+ double positive T cell density in locally advanced N2 non-small-cell lung carcinoma. Eur J Cancer 2020;135:221-9. [Crossref] [PubMed]
- Gataa I, Mezquita L, Rossoni C, et al. Tumour-infiltrating lymphocyte density is associated with favourable outcome in patients with advanced non-small cell lung cancer treated with immunotherapy. Eur J Cancer 2021;145:221-9. [Crossref] [PubMed]
- Banchereau R, Chitre AS, Scherl A, et al. Intratumoral CD103+ CD8+ T cells predict response to PD-L1 blockade. J Immunother Cancer 2021;9:e002231. [Crossref] [PubMed]
- Sumitomo R, Hirai T, Fujita M, et al. M2 tumor-associated macrophages promote tumor progression in non-small-cell lung cancer. Exp Ther Med 2019;18:4490-8. [Crossref] [PubMed]
- Edlund K, Madjar K, Mattsson JSM, et al. Prognostic Impact of Tumor Cell Programmed Death Ligand 1 Expression and Immune Cell Infiltration in NSCLC. J Thorac Oncol 2019;14:628-40. [Crossref] [PubMed]
- Lohr M, Edlund K, Botling J, et al. The prognostic relevance of tumour-infiltrating plasma cells and immunoglobulin kappa C indicates an important role of the humoral immune response in non-small cell lung cancer. Cancer Lett 2013;333:222-8. [Crossref] [PubMed]
- Yamauchi Y, Safi S, Blattner C, et al. Circulating and Tumor Myeloid-derived Suppressor Cells in Resectable Non-Small Cell Lung Cancer. Am J Respir Crit Care Med 2018;198:777-87. [Crossref] [PubMed]
- Backman M, Strell C, Lindberg A, et al. Spatial immunophenotyping of the tumour microenvironment in non-small cell lung cancer. Eur J Cancer 2023;185:40-52. [Crossref] [PubMed]
- Kobayashi K, Hernandez LD, Galán JE, et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 2002;110:191-202. [Crossref] [PubMed]
- Rhyasen GW, Starczynowski DT. IRAK signalling in cancer. Br J Cancer 2015;112:232-7. [Crossref] [PubMed]
- Jain A, Kaczanowska S, Davila E. IL-1 Receptor-Associated Kinase Signaling and Its Role in Inflammation, Cancer Progression, and Therapy Resistance. Front Immunol 2014;5:553. [Crossref] [PubMed]
- Standiford TJ, Kuick R, Bhan U, et al. TGF-β-induced IRAK-M expression in tumor-associated macrophages regulates lung tumor growth. Oncogene 2011;30:2475-84. [Crossref] [PubMed]
- del Fresno C, Otero K, Gómez-García L, et al. Tumor cells deactivate human monocytes by up-regulating IL-1 receptor associated kinase-M expression via CD44 and TLR4. J Immunol 2005;174:3032-40. [Crossref] [PubMed]
- Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012;12:253-68. [Crossref] [PubMed]
- Kesselring R, Glaesner J, Hiergeist A, et al. IRAK-M Expression in Tumor Cells Supports Colorectal Cancer Progression through Reduction of Antimicrobial Defense and Stabilization of STAT3. Cancer Cell 2016;29:684-96. [Crossref] [PubMed]
- Rothschild DE, Zhang Y, Diao N, et al. Enhanced Mucosal Defense and Reduced Tumor Burden in Mice with the Compromised Negative Regulator IRAK-M. EBioMedicine 2017;15:36-47. [Crossref] [PubMed]
- Wu X, Ouyang Y, Wang B, et al. Hypermethylation of the IRAK3-Activated MAPK Signaling Pathway to Promote the Development of Glioma. Cancer Manag Res 2020;12:7043-59. [Crossref] [PubMed]
- Geng D, Ciavattone N, Lasola JJ, et al. Induction of IRAK-M in melanoma induces caspase-3 dependent apoptosis by reducing TRAF6 and calpastatin levels. Commun Biol 2020;3:306. [Crossref] [PubMed]
- Saenger Y, Magidson J, Liaw B, et al. Blood mRNA expression profiling predicts survival in patients treated with tremelimumab. Clin Cancer Res 2014;20:3310-8. [Crossref] [PubMed]
- Caba O, Prados J, Ortiz R, et al. Transcriptional profiling of peripheral blood in pancreatic adenocarcinoma patients identifies diagnostic biomarkers. Dig Dis Sci 2014;59:2714-20. [Crossref] [PubMed]
- Angulo JC, Andrés G, Ashour N, et al. Development of Castration Resistant Prostate Cancer can be Predicted by a DNA Hypermethylation Profile. J Urol 2016;195:619-26. [Crossref] [PubMed]
- Kuo CC, Shih YL, Su HY, et al. Methylation of IRAK3 is a novel prognostic marker in hepatocellular carcinoma. World J Gastroenterol 2015;21:3960-9. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Győrffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharmacol 2024;181:362-74. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Han Y, Wang Y, Dong X, et al. TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res 2023;51:D1425-31. [Crossref] [PubMed]
- Kim N, Kim HK, Lee K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020;11:2285. [Crossref] [PubMed]
- Dost AFM, Moye AL, Vedaie M, et al. Organoids Model Transcriptional Hallmarks of Oncogenic KRAS Activation in Lung Epithelial Progenitor Cells. Cell Stem Cell 2020;27:663-678.e8. [Crossref] [PubMed]
- Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550-8. [Crossref] [PubMed]
- Fu J, Li K, Zhang W, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med 2020;12:21. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483:570-5. [Crossref] [PubMed]
- Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016;166:740-54. [Crossref] [PubMed]
- Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform 2021;22:bbab260. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. NCCN Guidelines® Insights: Non-Small Cell Lung Cancer, Version 2.2023. J Natl Compr Canc Netw 2023;21:340-50. [Crossref] [PubMed]
- Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 2012;30:2678-83. [Crossref] [PubMed]
- Zhao Y, Ge X, He J, et al. The prognostic value of tumor-infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: a systematic review and meta-analysis. World J Surg Oncol 2019;17:85. [Crossref] [PubMed]
- Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015;14:561-84. [Crossref] [PubMed]
- Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017;168:707-23. [Crossref] [PubMed]
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486-99. [Crossref] [PubMed]
- Muzio M, Ni J, Feng P, et al. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 1997;278:1612-5. [Crossref] [PubMed]
- Wesche H, Gao X, Li X, et al. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J Biol Chem 1999;274:19403-10. [Crossref] [PubMed]
- Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [Crossref] [PubMed]
- Norton JT, Hayashi T, Crain B, et al. Role of IL-1 receptor-associated kinase-M (IRAK-M) in priming of immune and inflammatory responses by nitrogen bisphosphonates. Proc Natl Acad Sci U S A 2011;108:11163-8. [Crossref] [PubMed]
- Balaci L, Spada MC, Olla N, et al. IRAK-M is involved in the pathogenesis of early-onset persistent asthma. Am J Hum Genet 2007;80:1103-14. [Crossref] [PubMed]
- Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol 2013;25:261-7. [Crossref] [PubMed]
- Watnick RS. The role of the tumor microenvironment in regulating angiogenesis. Cold Spring Harb Perspect Med 2012;2:a006676. [Crossref] [PubMed]
- Klebanoff CA, Gattinoni L, Restifo NP. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol Rev 2006;211:214-24. [Crossref] [PubMed]
- Schioppa T, Moore R, Thompson RG, et al. B regulatory cells and the tumor-promoting actions of TNF-α during squamous carcinogenesis. Proc Natl Acad Sci U S A 2011;108:10662-7. [Crossref] [PubMed]
- Olkhanud PB, Damdinsuren B, Bodogai M, et al. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4+ T cells to T-regulatory cells. Cancer Res 2011;71:3505-15. [Crossref] [PubMed]
- Franklin RA, Liao W, Sarkar A, et al. The cellular and molecular origin of tumor-associated macrophages. Science 2014;344:921-5. [Crossref] [PubMed]
- Shiga K, Hara M, Nagasaki T, et al. Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers (Basel) 2015;7:2443-58. [Crossref] [PubMed]
- Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol 2012;12:157-67. [Crossref] [PubMed]
- Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res 2019;79:4557-66. [Crossref] [PubMed]
- Hou J, Karin M, Sun B. Targeting cancer-promoting inflammation - have anti-inflammatory therapies come of age? Nat Rev Clin Oncol 2021;18:261-79. [Crossref] [PubMed]
- Xue Q, Wang Y, Zheng Q, et al. Prognostic value of tumor immune microenvironment factors in patients with stage I lung adenocarcinoma. Am J Cancer Res 2023;13:950-63. [PubMed]
- Tunalı G, Rúbies Bedós M, Nagarajan D, et al. IL-1 receptor-associated kinase-3 acts as an immune checkpoint in myeloid cells to limit cancer immunotherapy. J Clin Invest 2023;133:e161084. [Crossref] [PubMed]
- Turnis ME, Song XT, Bear A, et al. IRAK-M removal counteracts dendritic cell vaccine deficits in migration and longevity. J Immunol 2010;185:4223-32. [Crossref] [PubMed]
- Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat Rev Cancer 2021;21:345-59. [Crossref] [PubMed]
- Haslam A, Prasad V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for and Respond to Checkpoint Inhibitor Immunotherapy Drugs. JAMA Netw Open 2019;2:e192535. [Crossref] [PubMed]
- Garcia-Diaz A, Shin DS, Moreno BH, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 2017;19:1189-201. [Crossref] [PubMed]
- Spranger S, Spaapen RM, Zha Y, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 2013;5:200ra116. [Crossref] [PubMed]
- Knopf P, Stowbur D, Hoffmann SHL, et al. Acidosis-mediated increase in IFN-γ-induced PD-L1 expression on cancer cells as an immune escape mechanism in solid tumors. Mol Cancer 2023;22:207. [Crossref] [PubMed]
- Benci JL, Xu B, Qiu Y, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016;167:1540-1554.e12. [Crossref] [PubMed]
- Theivanthiran B, Evans KS, DeVito NC, et al. A tumor-intrinsic PD-L1/NLRP3 inflammasome signaling pathway drives resistance to anti-PD-1 immunotherapy. J Clin Invest 2020;130:2570-86. [Crossref] [PubMed]
- Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018;15:234-48. [Crossref] [PubMed]


