
TRIM27 revealing by tumor educated platelet RNA-sequencing, as a potential biomarker for malignant ground-glass opacities diagnosis mediates glycolysis of non-small cell lung cancer cells partially through HOXM1
Highlight box
Key findings
• In this study, a novel classifier based on tumor educated platelet (TEP)-RNA sequencing for distinguishing malignant from benign ground-glass opacities (GGOs) was developed and evaluated the diagnostic performance. Tripartite motif-containing 27 (TRIM27), as a potential biomarker, achieved area under the curve of 0.87 in the diagnosis of malignant GGOs, with 83.93% sensitivity, 78.79% specificity.
What is known and what is new?
• TRIM27 is high expression and acts as an oncogene in lung cancers, in vitro, TRIM27 promoted cell migration and invasion, which suggested that TRIM27 may be an important regulator of cell motility and metastasis.
• TRIM27 promoted glycolysis through regulates adenosine triphosphate, reactive oxygen species, and lactate acid production, glucose uptake, and lactate dehydrogenase activity by altering the activities of HK1/2, PKM1/2, LDHA, and GLUT1. Furtherly, HMOX1 was one of a downstream gene negatively correlated with TRIM27 and could be a partial target of TRIM27 mediating glycolysis in non-small cell lung cancer cells.
What is the implication, and what should change now?
• TRIM27 may be considered for clinical application as a diagnostic biomarker of malignant GGOs based on TEP RNA.
Introduction
The widespread use of low-dose computed tomography to screen for lung cancer has inevitably led to the detection of numerous small, asymptomatic lung nodules and ground-glass opacities (GGOs), which are defined as any hazy lung opacities that do not obscure the underlying bronchial structures or pulmonary vessels (1). GGOs may manifest with various clinical features (2). While some GGOs are benign, a considerable proportion are malignant. Strategies for managing these screening-detected lesions to exclude cancer and avoid “missed diagnoses” or “over-diagnoses” are imperative (1). Therefore, developing a noninvasive, accurate, and effective method for detecting benign and malignant GGOs could be tremendously beneficial in clinical practice.
Blood platelets, the second most abundant peripheral-blood cell type originating from megakaryocytes, have emerged as key players in systemic and local responses to tumor growth (3,4). During the bidirectional tumor-platelet interactions, platelets constantly absorb and enrich tumor-specific substances in the circulation, which can alter their RNA profiles thus termed “tumor educated platelet (TEP)” (3,5). Platelet RNA transcripts are derived from parental megakaryocytes during platelet origination (6). However, by stimulating signals released from tumor cells and the tumor microenvironment, platelets can be activated and induce specific splicing of pre-messenger RNA (pre-mRNA) to ingest circulating mRNA, thus providing unique and dynamic mRNA profiles with potential applicability in cancer diagnosis (7). Due to the regulatory mechanism of post-transcriptional splicing, the alterations of platelets mRNA profiles have been identified as tumor markers (5). In individuals with non-small cell lung cancer (NSCLC), TEP RNA profiles were reported to achieve accuracies of 81% and 88% in early-and late-stage cohorts, respectively, by particle-swarm optimization-enhanced algorithms, independent of age, smoking habits, and various inflammatory statuses (8). In this study, a classifier was constructed to detect malignant GGOs using the differentially expressed genes (DEGs) derived from TEP RNA-seq between benign and malignant GGOs, and reliable preliminary results were yielded.
Glycolysis is a vital factor that promotes cancer development (9). Compared with normal cells, rapidly proliferating cancer cells exhibit a higher rate of glucose consumption, and part of their glucose carbon is converted to lactate in a process called aerobic glycolysis, also known as “the Warburg effect” (10). The production of nucleic acids, lipids, and proteins is essential for successful replicative cell division (11). Glycolytic intermediates are crucial precursors involved in DNA, lipid, and protein syntheses (12), potentially explaining why cancer cells prefer glycolysis.
Through high-throughput quantitative polymerase chain reaction (HT-qPCR) verification of lung cancer and lung inflammation patients with GGOs as well as correlation analysis between the verified DEGs and key glycolytic genes, tripartite motif-containing 27 (TRIM27) emerges as a potential biomarker for distinguishing malignant from benign GGOs based on TEP RNA detection. TRIM27 belongs to the TRIM family (13,14) and acts as an oncogene in NSCLC (15,16). However, its role in NSCLC remains to be explored. In the present study, it demonstrates that TRIM27 correlated with higher motility and glycolysis, and its regulation towards glycolysis is partially through HMOX1 in NSCLC cells. Our findings provide a scientific basis for TRIM27 as an effective molecular marker for diagnosing malignant GGOs. We present this article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-157/rc).
Methods
Clinical samples and cell lines
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All clinical samples were obtained from The Second Xiangya Hospital of Central South University. This study was approved by the Medical Ethics Committee of The Second Xiangya Hospital of Central South University (No. 2020-ethical review-clinical study-107). Informed consent was obtained from all participants involved in the study. Whole-blood samples were collected from patients with GGOs regardless of whether they were benign or malignant. Blood was drawn into 10-mL ethylenediaminetetraacetic acid-coated anticoagulant tubes (BD Biosciences, Franklin Lakes, NJ, USA). Subsequently, based on the clinical diagnosis, patients were divided into malignant and inflammatory groups. Cancer and adjacent tissues were collected from 14 patients and stored at −80 ℃ until used.
The human lung fibroblast cell line MRC-5 (icellbioscience, iCell-h146, Shanghai, China) (17) and NSCLC cell lines A549 (Science&Well, SSW-1201, Changsha, China) and PC-9 (Science&Well, SSW-1202) and cultured in DMEM (Basal Media, L110KJ, Shanghai, China). NCI-H1975 cells (Science&Well, SSW-1205) were grown in RPMI-1640 medium (Basal Media, L210KJ, Shanghai, China). The cell lines were obtained from American Type Culture Collection (ATCC). All culture media were supplemented with 10% fetal bovine serum (FBS; Biological Industries, 04-001-1ACS, Kibbutz Beth Haemek, Israel) and 1% penicillin/streptomycin (Solarbio, P1400, Beijing, China). Cells were incubated at 37 ℃ with 5% CO2.
Blood processing and platelet isolation
Blood samples were processed within 48 h. Platelets were isolated from whole blood using two-step centrifugation. The first centrifugation step was implemented to separate platelet-rich plasma from erythrocytes and leukocytes by spinning for 20 min at 120 ×g and 4 ℃. Thereafter, the second centrifugation step was conducted to enrich platelet pellets by spinning for 20 min at 360 ×g and 4 ℃. The platelet pellets were carefully resuspended in 30 µL RNAlater® (Invitrogen, AM7024, Carlsbad, USA), incubated overnight at 4 ℃, and subsequently frozen at −80 ℃ until used.
RNA extraction and cDNA synthesis
Platelet RNA was extracted in batches, frozen platelet pellets were thawed on ice, and total RNA was isolated using the QIAamp® RNA Blood Mini Kit (Qiagen, 52304, Dusseldorf, Germany). Extraction from cells and tissues was conducted using an RNA extraction kit (Tiangen, DP419, Beijing, China), and 200 ng of RNA was reverse transcribed to cDNA using commercial SMARTScribe™ Reverse Transcriptase (TaKaRa, 639538, Tokyo, Japan).
Library preparation and sequencing of TEP RNA
Platelet RNA-seq library preparation was performed according to a standardized process (18). cDNA (200 ng) was amplified using the KAPA HiFi™ HotStart ReadyMix PCR Kit (Roche, 09420398001, Basel, Switzerland). To label platelet cDNA for sequencing, all amplified platelet cDNA was subjected to nucleic-acid shearing and labeled with the Tn5 adapter using Tn5 Transposase (MDTK Bio, MD3001-100, Beijing, China). Subsequently, the bead-purified product processed by Tn5 Transposase was labeled with a single-index barcode using the KAPA2G Robust PCR Kit (Merck, KK5023, Darmstadt, Germany). All the purification steps were performed using AMPure XP beads (Beckman Coulter, A63882, Miami, USA). Commercial kits were used according to the respective manufacturers’ instructions. The final platelet RNA libraries were quantified using the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and sequenced on the Illumina NovaSeq sequencing platform (Illumina, San Diego, CA, USA) in 150-bp paired-end mode.
Raw platelet RNA-seq data encoded in FASTQ files were analyzed as described previously (7). Mapping, summarization, and normalization of RNA-seq reads were also performed as described previously (8,18). The full cohort comprised 81 patients who were randomly divided into training and test sets in a 7:3 ratio. DEGs analysis was conducted in the training set using the R package DESeq2. Diagnostic analysis for benign and malignant GGOs was performed using the random forest algorithm, and the diagnostic results were utilized to process receiver operating characteristic (ROC) analysis. ROC analyses of the training set, test set, and full cohort were performed using the R package pROC (version 1.18.0), and the results were presented as ROC curves and summarized using area under the curve (AUC) values with 95% confidence intervals (CIs).
RNA-seq of TRIM27 loss-of-function cell models
To explore the pathways downstream of TRIM27 that exert its biological functions, RNA-seq was performed in PC-9 cell models with or without TRIM27 knockdown (KD) (two groups: si-NC and si-TRIM27, two parallel samples each). DEGs with |log2 fold change (FC)| >1.0 and adjusted P values <0.05 were considered to have significant differences between groups.
HT-qPCR and real-time PCR assays
HT-qPCR analysis was performed using the WaferGen Real-Time PCR Cycler (WaferGen Biosystems, Fremont, USA) according to the manufacturer’s instruction (final 10 ng/µL cDNA, 250 nM primer, and 1× TB Green® Premix Ex Taq™ (Tli RNaseH Plus; TaKaRa, RR420A) dispensed using the WaferGen SmartChip™ Multisample Nanodispenser. HT-qPCR analysis was conducted under cycling conditions of 10 min at 95 ℃ and subsequently 40 cycles of 30 s at 95 ℃ and 30 s at 60 ℃, followed by a melting curve. Conventional real-time PCR analysis was performed according to the manufacturer’s protocol: cDNA was mixed with SuperReal PreMix Plus (SYBR Green) Kits (Tiangen, FP205-02) and each primer pair to produce a 20-µL reaction mixture. The assays were conducted using an ABI 7500 Real-Time PCR System (Applied Biosystems). The primer-pair sequences are listed in Table S1, and β-actin was used as an internal control. All reactions were done in triplicates, the average 2−ΔΔCt was used in data analysis.
Cell transfection
Cell transfection was performed using Lipofectamine™ 3000 Transfection Reagent (Invitrogen, L3000-015) following the manufacturer’s instructions. A549 cells were transfected with TRIM27-pcDNA3.1(+) (YouBio, Changsha, China) to generate a TRIM27 gain-of-function cell model. PC-9 cells were used to construct a loss-of-function cell model of TRIM27 by transfecting small interfering RNA (siRNA) synthesized from GENERAL BIOL (Chuzhou, Anhui, China). The sequences of the target siRNAs are shown in Table S2. The siRNA sequence of HMOX1 was synthesized as described in reference (19), and plasmid of HMOX1-pcDNA3.1(+) was synthesized by YouBio.
Transwell assays
Invasion and migration assays were performed using Transwell® Permeable Supports (6.5-mm inserts; Corning, NY, USA) with 8.0-µm polycarbonate membrane. The cells transfected for 24 h were resuspended in serum-free DMEM. The upper chambers of the Transwell inserts were each seeded with 2×104 cells in 300 µL serum-free DMEM, and the cells were precoated with or without Matrigel™ (BD Biosciences, 356234). The lower chambers each had 500 µL DMEM containing 15% FBS added to them. After 24 h of incubation, the cells in the lower chambers were fixed with 4% paraformaldehyde (Beyotime, P0099, Shanghai, China) for 20 min and stained with 0.1% crystal violet solution (Solarbio, C8470) for 15 min. The cells that did not cross the membrane were removed using a cotton swab. Cells were photographed using a Motic AE31 microscope (Motic, Xiamen, China) and counted in three random fields.
Cell proliferation assays
The cells were resuspended and added to 96-well plates at a density of 5×103 cells/well for cell proliferation assays. Cell Counting Kit-8 (CCK-8; Beyotime, C0037) assays were performed according to the manufacturer’s instructions. At 0, 24, and 48 h, the CCK-8 assays were performed using an ELX-808 Microplate Reader (BioTek, Winooski, Vermont, USA) at 450 nm.
The YF® Click-iT 5-ethynyl-2'-deoxyuridine (EdU) Kit (BioScience, C6017M, Shanghai, China) was used to conduct EdU assays. Cells were incubated for 2 h with 100 µL of 50 µM EdU buffer, and positive cells were visualized using a Motic AE31 microscope.
Cell colony formation assays
Six hundred transfected cells were seeded in a 60-mm culture dish in triplicate. For the 14-day incubation, the cells were fixed with 1 mL of 4% paraformaldehyde for 15 min and stained with crystal violet solution for 10 min. After washing three times with phosphate-buffered saline, cell colonies were counted.
Apoptosis assays
Apoptosis assays were conducted using the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, 556547). After 48 h of transfection, the cells were harvested and washed twice with cold Dulbecco’s phosphate-buffered saline. Thereafter, cells were resuspended using 100 µL 1× binding buffer and stained with FITC Annexin V and propidium iodide. After incubation for 15 min, 400 µL 1× binding buffer was added. Finally, apoptosis was analyzed using flow cytometry.
Adenosine triphosphate (ATP) production, lactate dehydrogenase (LDH) activity, and lactate acid (LD) production assays
The model cells were resuspended and prepared for assays. ATP production assay was performed using an ATP Luminescence kit (UElandy, A6103S, Suzhou, China) following the manufacturer’s instructions. LD production and LDH activity assays were performed using LD (JianchengBio, A019-2-1, Nanjing, China) and LDH (JianchengBio, A020-2) assay kits according to the manufacturer’s instructions.
Glucose uptake and reactive oxygen species (ROS) production assays
For the glucose uptake assay, the culture medium was replaced with glucose- and serum-free DMEM, and the cells were incubated for 6 h. Subsequently, complete media with 120 µM 2-NBD-glucose (2-NBDG) (Invitrogen, N13195) were used to culture cells overnight. ROS production assay was performed using an ROS Assay Kit (Beyotime, S0033S) according to the manufacturer’s instructions. Finally, fluorescence levels were detected using flow cytometry, which was used to measure intracellular glucose uptake and ROS production.
Western blot
Total protein was extracted from the cells or tissue using radioimmunoprecipitation assay lysis buffer (TDY Biotech, WB0009, Beijing, China) containing phenylmethylsulfonyl fluoride and a protease inhibitor cocktail. Thereafter, the supernatants were collected from the cell lysates by centrifuging at 12,000 rpm for 20 min at 4 ℃. Protein concentrations were measured using a bicinchoninic acid (BCA) protein assay kit (TDY Biotech, WB0028) according to the manufacturer’s protocol. Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Subsequently, the membranes were respectively incubated with anti-TRIM27, anti-HK-1/2, anti-PKM, anti-LDHA, anti-GLUT1, anti-HMOX1 and anti-β-actin overnight at 4 ℃ and then incubated with the respective secondary antibodies. The antibodies used in this study are list in Table S3. Protein signals were detected using an ECL chemiluminescent substrate kit (NEN, Boston, MBS8305415, MA, USA). The densities of each band were quantified by the Image J program.
Double immunofluorescence labeling assays
The cells were seeded on slides overnight. After 48 h of transfection, the cell slides were fixed with 4% formaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 20 min, and then blocked with FBS for 1 h at room temperature. The cell slides were incubated with anti-HMOX1 and anti-TRIM27 overnight at 4 ℃, washed and further incubated with goat anti-Mouse IgG H&L (Cy3) (Ab97035, Abcam, Cambridge, UK) and goat anti-Rabbit IgG H&L (FITC) (Ab6717, Abcam) at room temperature in the dark for 30 min. The cell slides were observed and photographed under panoramic scanning system (Pannorramic MIDI, Budapest, Hungary). The fluorescence densities of three random views were quantified by the Image J software.
Statistical analysis
Data were statistically analyzed using GraphPad Prism (version 7.00, GraphPad Software, San Diego, USA). Two-tailed Student’s t-test and one-way analysis of variance were used to compare two or more groups. Statistical significance was set at P<0.05.
Results
Flow chart describing the study design for identifying malignant GGOs by platelets RNA profiles
Whole-blood samples were collected from 103 patients with malignant (n=68) and inflammatory (n=35) GGOs recruited from The Second Xiangya Hospital of Central South University from November 2020 to May 2021. Finally, 81 participants, comprising 59 and 22 with malignant and benign GGOs, respectively, were enrolled. Their basic characteristics are shown in Table 1. Among the 59 malignant GGOs patients, 57 patients were diagnosed lung adenocarcinoma (LUAC), one patient was diagnosed lung squamous cell carcinoma (LUSC) and one patient was unclassified. The schematic of the trial design for identifying malignant and benign GGOs based on platelet RNA profiles is shown in Figure 1.
Table 1
| Characteristics | Malignant GGO-lung cancer (n=59) | Benign GGO-inflammation (n=22) |
|---|---|---|
| Type | ||
| LUAC | 57 | – |
| LUSC | 1 | – |
| Unclassified | 1 | – |
| Gender | ||
| Male | 25 | 16 |
| Female | 34 | 6 |
| Age, years | ||
| Median | 63 | 55 |
| IQR | 18.25 | 15 |
| Min–max | 24–91 | 30–73 |
| Smoking | ||
| Current | 11 | 7 |
| Former | 0 | 1 |
| Never | 48 | 14 |
GGO, ground-glass opacity; LUAC, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; IQR, interquartile range.

Analysis of platelets RNA-seq and malignant GGO diagnosis
All TEP RNA-seq data were deposited in the National Center for Biotechnology Information Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) under project number PRJNA 1002810. The distribution of gene expression was uniform across the malignant and inflammatory cohort samples according to raw data preprocessing (Figure 2A), and the processed data were used for subsequent analysis. DEG analysis was performed using the training set, which included 38 malignant and 18 inflammatory samples. A total of 1,647 DEGs were identified (|log2FC| >1, adjusted P values <0.05), of which 674 genes were upregulated, and 973 were significantly downregulated (Figure 2B). Subsequently, cluster analysis was performed, as shown in Figure 2C, and the DEGs were well clustered. The top 30 upregulated and downregulated DEGs were selected for cluster analysis, which also exhibited good clustering (Figure 2D). To evaluate the diagnostic power of the TEP RNA profiles in diagnosing malignant GGOs, DEGs were calculated using the random forest algorithm. For the 1,647 DEGs, ROC analysis was performed, and the AUC of the training set, test set, and total samples were 0.906 (95% CI: 0.791–1.000), 0.881 (95% CI: 0.690–1.000), and 0.990 (95% CI: 0.972–1.000). Classification sensitivity, specificity, and accuracy were 100%, 90.91%, and 97.53%, respectively (Figure 2E).

Regarding the diagnostic power of the 60 DEGs, which comprised the top 30 upregulated and top 30 downregulated DEGs, the AUCs of the training set, test set, and total samples were 0.927 (95% CI: 0.826–1.000), 0.833 (95% CI: 0.579–1.000), and 0.986 (95% CI: 0.962–1.000), respectively. Classification sensitivity, specificity, and accuracy were 98.31%, 90.91%, and 96.30%, respectively (Figure 2F). These results suggest that the TEP RNA profiles displayed acceptable diagnostic power with reliable sensitivity and specificity in diagnosing malignant GGOs. In terms of the most significant 60 DEGs, the diagnostic efficiency was reliable.
Additionally, the Gene Set Enrichment Analysis (GSEA) was implemented, which results revealed the malignant GGOs group was significantly enriched in glycolysis and gluconeogenesis Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway which ranked first (Figure S1). The related original data was presented in the table available at https://cdn.amegroups.cn/static/public/tlcr-24-157-1.xlsx.
HT-qPCR verification of the 23 selected DEGs in platelet samples
HT-qPCR assays are typically used to verify sequencing results after RNA-seq. In this study, 23 DEGs selected from the top 60 DEGs were further locked into our follow-up research; however, their molecular functions had not been explored in the field of lung cancer, according to a PubMed consultation of references at the Q1–Q2 partition level. The 23 DEGs comprised 15 upregulated (ELP3, GIMAP6, GIMAP8, HSPA8, MCM3, NLRC4, PIK3AP1, PPWD1, PSEN1, RASSF2, RNASEL, SF3B3, SLFN5, SPTBN1, and TRIM27) and eight downregulated (ATP5MD, COX6B1, EMP3, FTL, HBE1, POLR2L, POMP, and SF3B6) DEGs in the malignant cohort. In order to verify the mRNA level of 23 DEGs, 56 malignant and 66 inflammatory samples was collected to perform HT-qPCR. The results demonstrated that the mRNA levels of 17 DEGs were consistent with the RNA-seq results, whereas those of six DEGs were inconsistent. The 17 consistent DEGs included 15 upregulated (ELP3, GIMAP6, GIMAP8, HSPA8, MCM3, NLRC4, PIK3AP1, PPWD1, PSEN1, RASSF2, RNASEL, SF3B3, SLFN5, SPTBN1, and TRIM27) and two downregulated (ATP5MD and HBE1) DEGs in the cancer groups compared with those in the inflammatory groups (P<0.05; Figure 3). The six inconsistent DEGs (COX6B1, EMP3, FTL, POLR2L, POMP, and SF3B6) were not markedly different in the cancer samples compared with those in the inflammation samples (Figure S2). In brief, HT-qPCR verification suggested that these 17 DEGs may have further significance in lung cancer progression.
Screening of glycolysis correlative genes from 17 DEGs
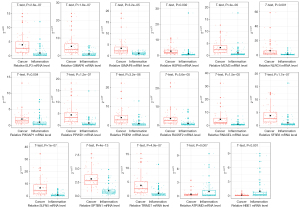
Glycolysis is an important feature of tumor development and progression (20). Some of the intergroup DEGs derived from TEP RNA-seq of the benign and malignant GGOs were speculated to have been potentially related to glycolysis regulation. A previous study selected 22 genes (SLC2A1, HK1, HK2, HK3, GPI, PFKL, PFKM, PFKP, ALDOA, ALDOB, ALDOC, TPI1, GAPDH, PGK1, PGAM1, PGAM4, ENO1, ENO2, ENO3, PKLR, PKM, and LDHA) whose expression signatures belonged to the core glycolytic pathway (9). Subsequently, a correlation analysis was performed between the 17 previously verified DEGs and 22 glycolysis core pathway genes mentioned above, using 59 platelet RNA-seq data from a malignant cohort. Most of the 15 upregulated DEGs were positively correlated with SLC2A1, HK2, HK3, and LDHA and negatively correlated with HK1, GPI, PFKL, PFKM, PFKP, ALDOA, TPI1, GAPDH, PGK1, PGAM1, ENO1, ENO2, and PKM, while the two downregulated DEGs (ATP5MD and HBE1) exhibited an opposite trend (Figure 4A).

In addition, seven DEGs (GIMAP8, HSPA8, MCM3, PIK3AP1, RNASEL, SF3B3, and TRIM27) considered to be relatively strongly associated with glycolysis were selected (|Pearson’s correlation| >0.7, P<0.05), and their relative mRNA levels were measured in 10 pairs of NSCLC and matched adjacent tissues (Figure 4B-4H). Real-time PCR assays revealed that the mRNA levels of GIMAP8 and PIK3AP1 were significantly decreased in the cancer group compared with those in the adjacent-tissue group, exhibiting inconsistency with the mRNA levels in platelets. Only TRIM27 mRNA levels were markedly increased in cancer tissues, and consistent with its expression in platelets. Previous studies have demonstrated that TRIM27 is highly expressed in NSCLC (15,16). Moreover, the average 2−ΔΔCt of TRIM27 in HT-qPCR assays was performed in ROC analysis to obtain the diagnostic performance of a single TRIM27 biomarker. The ROC curves demonstrated an AUC of 0.87, 83.93% sensitivity, 78.79% specificity, 81.15% accuracy, 77.05% positive predictive value (PPV) and 85.25% negative predictive value (NPV) (Figures 4I). Thereby, we will next investigate the functional role of TRIM27 in NSCLC cells.
TRIM27 regulated cell migration and invasion in NSCLC cells
First, TRIM27 expression was examined in NSCLC cell lines (NCI-H1975, A549, and PC-9) and the human lung fibroblast cell line MRC5. TRIM27 was found to be evidently enhanced in NSCLC cells compared with that in MRC5 cells. In addition, among the three NSCLC cell lines, the endogenous expression of TRIM27 was relatively lower in A549 cells and highest in PC9 cells (Figure 5A). A549 cells were transfected with TRIM27-pcDNA3.1(+) to construct a TRIM27 gain-of-function cell model, and a TRIM27 loss-of-function cell model was constructed in PC-9 cells by transfection with three alternative TRIM27 siRNAs, among which si-TRIM27-1259 exhibited the strongest inhibition efficiency and was used for subsequent assays (Figure 5B). Meanwhile, TRIM27 protein expression in different cell lines was verified by western blotting (Figure 5C). The functional role of TRIM27 was also investigated in vitro. Neither silencing nor overexpression of TRIM27 resulted in significant differences in cell proliferation in the CCK8 and EdU assays (Figure 5D,5E). Correspondingly, as regards A549 or PC9 cells, no significant difference in clonality was noted between groups based on the cell colony formation assays (Figure 5F). In the cell apoptosis assays, TRIM27 overexpression did not alter the apoptotic rate of A549 cells; nevertheless, the apoptotic rate was significantly increased in TRIM27-KD PC-9 cells (Figure 5G). More importantly, TRIM27 markedly regulated cell migration and invasion. The migration and invasion of TRIM27-overexpressing A549 cells (oe-TRIM27) were significantly elevated compared with those of oe-NC, whereas the migration and invasion of PC-9 cells with TRIM27 KD (si-TRIM27) were obviously decreased compared with those of si-NC (Figure 5H). These data suggest that TRIM27 plays an important role in NSCLC cell motility.

TRIM27 promoted glycolysis in NSCLC cells
Based on correlation analysis, TRIM27 was strongly correlated with 22 core glycolytic pathway genes. Moreover, TRIM27 accelerates glucose uptake in esophageal squamous cell carcinoma (ESCC) cells by upregulating GLUT1 and HK2 to promote glycolysis (21). In this study, the relative mRNA expression levels of seven key glycolytic genes (PKM1, PKM2, PFKL, LDHA, HK1, HK2, and GLUT1) were detected using real-time PCR in A549 and PC-9 cell models. The results revealed that the relative mRNA levels of these seven genes were not significantly different between the oe-NC and oe-TRIM27 groups in A549 cells; however, the relative mRNA levels of the five key glycolysis genes (PKM1, PKM2, LDHA, HK1, and HK2) were significantly reduced in si-TRIM27 PC-9 cells compared with those in si-NC (Figure 6A). Western blot was performed to verify the protein levels of PKM1, PKM2, LDHA, HK1, HK2, and GLUT1. The protein levels of HK1/2, PKM1/2, LDHA, and GLUT1 decreased when TRIM27 was silenced in PC-9 cells. The opposite results were observed in TRIM27-overexpressing A549 cells (Figure 6B). Moreover, the protein expression of HK1/2, PKM1/2, LDHA, GLUT1 was detected in the seven paired NSCLC and adjacent tissues. Compared with the adjacent tissue group, the protein expression of the above genes was obviously elevated in the cancer tissue group (Figure S3). Subsequently, we explored the effects of TRIM27 on glycolysis in A549 and PC-9 cells. Glucose uptake, ROS, ATP and LD production, and LDH activity are the main features of glycolysis (22-24). Overexpressed TRIM27 markedly enhanced glucose uptake in A549 cells (vs. oe-NC), whereas a striking decrease was observed in TRIM27-KD PC-9 cells (vs. si-NC) (Figure 6C). ROS production was reduced in TRIM27-overexpressing A549 cells and elevated in TRIM27-KD PC-9 cells (Figure 6D). Meanwhile, overexpressed TRIM27 significantly accelerated ATP and LD production, whereas inhibiting TRIM27 markedly decreased ATP and LD production (Figure 6E,6F). However, LDH activity did not improve when TRIM27 was overexpressed but was markedly attenuated after TRIM27 silencing (Figure 6G). These results demonstrate that TRIM27 regulates ATP, ROS, and LD production, glucose uptake, and LDH activity by altering the activities of HK1/2, PKM1/2, LDHA, and GLUT1.

TRIM27/HMOX1 axis regulated glycolysis in NSCLC cells
To further explore the downstream targets and pathways of TRIM27 in NSCLC cells, RNA-seq was performed using TRIM27-KD PC-9 cells. In total, 162 upregulated and 116 downregulated DEGs were identified when TRIM27 was knocked down in PC-9 cells (vs. si-NC) (Figure 7A). Subsequently, the top 10 upregulated and downregulated DEGs were screened for cluster analysis, as shown in the heatmap (Figure 7B). In upregulated DEGs with TRIM27 KD, EPHA6 is considered a putative tumor suppressor in glioblastoma multiforme that promotes apoptosis (25). HMOX1 encodes heme oxygenase 1 which is involved in mediating oxidative stress, therapeutic resistance, hypoxia, and ferroptosis in lung cancer (26-29), and has been shown to inhibit cell growth and metastasis in lung mucoepidermoid carcinoma (30). High FKBPL levels inhibit breast cancer cell growth and sensitize cells to endocrine therapy (31,32). In downregulated DEGs with TRIM27 KD, IARS2 silencing inhibited proliferation, promoted apoptosis in NSCLC cells, and inhibited tumor growth in vivo (33). Upregulated DZIP1 has been found to directly promote the invasion and metastasis of gastric cancer cells and exhibit correlation with the immunosuppressive microenvironment of gastric cancer (34). Certain studies have reported that SLC39A6 downregulation suppresses ESCC cell growth and invasive ability and induces apoptosis (35,36). These results indicate that TRIM27 potentially promotes tumorigenesis by regulating multiple targets, including other top DEGs that lack tumorigenesis-related studies.
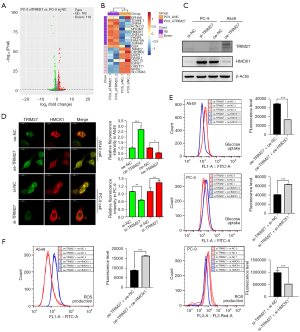
Among the above genes, HMOX1 caught our attention which was furtherly explored whether it was a downstream gene of TRIM27. Based on RNA-seq analysis in TRIM27-KD PC-9 cells, HMOX1 was upregulated with TRIM27 KD. To verify this result, western blot and double immunofluorescence staining were conducted in cell models. As anticipated, the protein expression of HMOX1 was increased in TRIM27-KD PC-9 cells and decreased in TRIM27-overexpressing A549 cells (Figure 7C). In addition, the protein expression of TRIM27 and HMOX1 was detected in the seven paired NSCLC and adjacent tissue. Compared with the adjacent tissue group, the protein expression of TRIM27 was obviously increased in the cancer tissue group while the protein expression of HMOX1 was obviously reduced in the cancer tissue group comparing to the adjacent tissue group (Figure S4). Moreover, in TRIM27 overexpressed A549 cells, the fluorescence density of HMOX1 (red) was reduced when TRIM27 (green) was overexpressed. While, in TRIM27-KD PC-9 cells, the red fluorescence density of HMOX1 was enhanced obviously when TRIM27 was reduced (Figure 7D). Furtherly, the glucose uptake and ROS production were investigated in TRIM27/HMOX1 dual cell models. In TRIM27-overexpressing A549 cells, HMOX1 overexpression decreased glucose uptake and increased ROS production obviously when compared with TRIM27 overexpressed A549 cells without HMOX1 overexpression (oe-TRIM27 + oe-NC). In the other hand, in TRIM27-KD PC-9 cells, inhibiting HMOX1 elevated glucose uptake and reduced ROS production (Figure 7E,7F). These results suggested that HMOX1 was one of a downstream gene negatively correlated with TRIM27 and could be a partial target of TRIM27 mediating glycolysis in NSCLC cells.
Discussion
In recent years, several studies have focused on TEP and their applications in tumor diagnosis. Best et al. provided a valuable platform for companion diagnostics in pan-cancer and multiclass cancers by applying TEP mRNA profiles (7). Thereafter, a TEP RNA biomarker panel that could distinguish patients with NSCLC from individuals with various non-cancerous inflammatory conditions was identified (8). Recently, based on TEP-derived RNA profiles, a pan-cancer tumor site-of-origin classifier was developed to identify the tumor site of origin (37). Usually, the biomarkers for cancer diagnosis achieved from sequencing and data analysis are subjected to further qPCR verification and ROC analysis to obtain each potential biomarker’s AUC, sensitivity, and specificity values, which are beneficial for screening optimum targets (38). Recent studies based on TEP RNA-seq have been dedicated to developing a novel platform containing an abundance of potential mRNA biomarkers and surrogate RNA profiles that could be applied to cancer detection with superior sensitivity and specificity (8,18,37,39), and certain potential biomarkers and their diagnostic performance warrant further exploration. In this study, we developed a noninvasive diagnostic method based on TEP RNA-seq to detect benign and malignant GGOs with high diagnostic performance. The least absolute shrinkage and selection operator regression algorithm and support vector machines are common machine-learning algorithms that are typically utilized to obtain candidate targets (7,40). For candidate targets, verifying multiple cohorts via the qPCR assay and analyzing their availability using ROC curves are necessary to achieve a diagnostic panel containing a few outstanding biomarkers (41). Unfortunately, a diagnostic panel of clinical translational value was not developed in the present study, and such a panel will be screened and verified in a considerably larger sample in future work.
Although, there are many biomarkers founded that can be used to diagnose the cancers, few studies have proved the expression of biomarker genes in platelets can affect tumor activity. Platelets, as circulating anucleated cells, originate from megakaryocytes in bone marrow and lung niches (42). The majority of platelets remain quiescent during their 7- to 10-day lifespan (43), the relatively short lifespan makes stable platelets culture a difficult problem to perform. Despite lacking nuclei, platelets retain cytoplasmatic RNA derived from megakaryocytes and may translate small amounts of mRNAs and process miRNAs (44). Human pluripotent stem cells, including human embryonic stem cells and induced pluripotent stem cells, can induce platelets or their precursor megakaryocytes (45). Megakaryocytes can be transfected miRNA, and the effect of miRNA on platelets also can be observed (46). However, in the absence of relevant technology and experience, it was failed to implement the relative research under our current situation.
In terms of evaluating the survival ability of NSCLC cells, neither overexpression in A549 cells nor interference with TRIM27 in PC-9 cells exhibited significant regulatory differences in the CCK8, EdU and clone formation assays, implying that TRIM27 does not play a biological role in promoting survival. Subsequently, we found that apoptosis increased in TRIM27-KD PC-9 cells, exhibiting concordance with previous findings in ESCC and non-triple-negative breast cancer (21,47). However, significant inter-group apoptotic differences remained unnoticed in the A549 model cells (oe-TRIM27 vs. oe-NC). This was speculated to potentially emanate from the extremely low background apoptosis in A549 cell, observing the infeasibility of a further significant decrease in apoptosis through the overexpression of the oncogenic factor TRIM27. However, our results revealed a noticeable promotion of TRIM27 overexpression in A549 cells and obvious inhibition of TRIM27-KD in PC-9 cells on migration and invasion, suggesting that TRIM27 may be an important regulator of cell motility and metastasis, displaying consistency with a previous study that demonstrated the same overexpression in NSCLC cell H1975 (16).
Previous report has demonstrated that TRIM27 enhances the glycolysis activity by increasing GLUT1 and HK2 in ESCC (21). Our study revealed that TRIM27 elevated glycolysis activity not only through upregulating GLUT1 and HK1/2 but also through increasing PKM1/2 and LDHA in NSCLC cells. LDH is a pivotal enzyme in driving glycolysis and is responsible for the final step in the conversion of pyruvate to lactate, thus playing a significant role in cancer progression, survival, invasion, angiogenesis, and metastasis via tumor glycolysis (48,49). Lung cancer cells exhibit a metabolic phenotype characterized by increased glucose uptake and glycolytic activity to support rapid growth (49-51), which is necessary for high LDH activity to ensure the smooth completion of glycolysis. In the present study, LDH activity was found to decrease in TRIM27-KD PC-9 cells but did not evidently increase in TRIM27-overexpressing A549 cells, thus indicating that the LDH activity of PC9 cells is potentially reduced by TRIM27 downregulation. However, increasing the LDH activity of A549 cells further with TRIM27 overexpression was difficult because its background LDH activity is already high.
Glycolysis is a typical metabolic rearrangement that provides abundant ATP and minimum ROS during the proliferation, migration, and invasion of tumor cells (52). Oncogenes facilitate cell proliferation, invasion, and metastasis by regulating glycolysis (53). Decreased LDH activity and lactate production have been reported to reduce cell motility in breast cancer (54). In this study, TRIM27 expression were correlated with the migration, invasion and glycolysis positively of NSCLC cells. Further, through RNA-seq of TRIM27 loss-of-function PC9 cells, the upregulated HMOX1 was found. HMOX1 was reported to significantly impair aerobic glycolysis in prostate cancer (55). In this research, HMOX1 overexpression also reduced glycolysis which was presented through the obvious decreasement of glucose uptake and a significant enhancement of ROS production in NSCLC. However, whether TRIM27 facilitated NSCLC cell apoptosis, migration, and invasion by regulating glycolysis and whether HMOX1 was involved have not yet been clarified. The glucose derivative 2-deoxy-d-glucose is an efficient tool for inhibiting glycolysis (56), which usually uses in the research of glycolysis. Therefore, whether glycolysis inhibition by molecular and pharmacological means can alter the apoptosis and motility of TRIM27 gain-of-function NSCLC cells will determine the possibility of a causal relationship between the glycolytic and phenotypic regulation of TRIM27. This aspect will be explored in future studies.
Conclusions
In conclusion, in our study, it was found that targeting TEP-derived RNA biomarkers could be a potentially promising diagnostic approach to distinguish malignant GGOs. TRIM27 was found as a biomarker emerged through TEP RNA-seq, which accelerated NSCLC cell migration and invasion and regulated glycolysis partially through HMOX1. Our work provided a scientific support for TRIM27 as a diagnostic biomarker for malignant GGO diagnosis to some extent for the first time.
Acknowledgments
Funding: The present study was supported by the National Natural Science Foundation of China (No. 81972638), Natural Science Foundation of Hunan Province (Nos. 2021JJ40838 and 2023JJ40825), Science and Technology Program of Changsha, China (No. kh2201053), Changsha Municipal Natural Science Foundation (No. kq2208333) and Clinical Nursing Research Fund Project of The Second Xiangya Hospital of Central South University (No. 2021-HLKY-01).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-157/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-157/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-157/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-157/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of The Second Xiangya Hospital of Central South University (No. 2020-ethical review-clinical study-107). Informed consent was obtained from all participants involved in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sihoe ADL. Should sublobar resection be offered for screening-detected lung nodules? Transl Lung Cancer Res 2021;10:2418-26. [Crossref] [PubMed]
- Kobayashi Y, Mitsudomi T. Management of ground-glass opacities: should all pulmonary lesions with ground-glass opacity be surgically resected? Transl Lung Cancer Res 2013;2:354-63. [PubMed]
- Antunes-Ferreira M, Koppers-Lalic D, Würdinger T. Circulating platelets as liquid biopsy sources for cancer detection. Mol Oncol 2021;15:1727-43. [Crossref] [PubMed]
- In 't Veld SGJG, Wurdinger T. Tumor-educated platelets. Blood 2019;133:2359-64. [Crossref] [PubMed]
- Ding S, Dong X, Song X. Tumor educated platelet: the novel BioSource for cancer detection. Cancer Cell Int 2023;23:91. [Crossref] [PubMed]
- Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. [Crossref] [PubMed]
- Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. [Crossref] [PubMed]
- Best MG, Sol N, In 't Veld SGJG, et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell 2017;32:238-252.e9.
- Wei J, Huang K, Chen Z, et al. Characterization of Glycolysis-Associated Molecules in the Tumor Microenvironment Revealed by Pan-Cancer Tissues and Lung Cancer Single Cell Data. Cancers (Basel) 2020;12:1788. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol 2019;20:436-50. [Crossref] [PubMed]
- Birts CN, Banerjee A, Darley M, et al. p53 is regulated by aerobic glycolysis in cancer cells by the CtBP family of NADH-dependent transcriptional regulators. Sci Signal 2020;13:eaau9529. [Crossref] [PubMed]
- Wang J, Zhao D, Lei Z, et al. TRIM27 maintains gut homeostasis by promoting intestinal stem cell self-renewal. Cell Mol Immunol 2023;20:158-74. [Crossref] [PubMed]
- Jaworska AM, Wlodarczyk NA, Mackiewicz A, et al. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 2020;38:165-73. [Crossref] [PubMed]
- Ji X, Lin L, Shen S, et al. Epigenetic-smoking interaction reveals histologically heterogeneous effects of TRIM27 DNA methylation on overall survival among early-stage NSCLC patients. Mol Oncol 2020;14:2759-74. [Crossref] [PubMed]
- Liu S, Tian Y, Zheng Y, et al. TRIM27 acts as an oncogene and regulates cell proliferation and metastasis in non-small cell lung cancer through SIX3-β-catenin signaling. Aging 2020;12:25564-80. [Crossref] [PubMed]
- Kim JY, An MJ, Shin GS, et al. Mercury Chloride but Not Lead Acetate Causes Apoptotic Cell Death in Human Lung Fibroblast MRC5 Cells via Regulation of Cell Cycle Progression. Int J Mol Sci 2021;22:2494. [Crossref] [PubMed]
- Best MG, In 't Veld SGJG, Sol N, et al. RNA sequencing and swarm intelligence-enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat Protoc 2019;14:1206-34.
- Rao Z, Zhang N, Xu N, et al. 1,25-Dihydroxyvitamin D Inhibits LPS-Induced High-Mobility Group Box 1 (HMGB1) Secretion via Targeting the NF-E2-Related Factor 2-Hemeoxygenase-1-HMGB1 Pathway in Macrophages. Front Immunol 2017;8:1308. [Crossref] [PubMed]
- Cui J, Shi M, Xie D, et al. FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin Cancer Res 2014;20:2595-606. [Crossref] [PubMed]
- Ma L, Yao N, Chen P, et al. TRIM27 promotes the development of esophagus cancer via regulating PTEN/AKT signaling pathway. Cancer Cell Int 2019;19:283. [Crossref] [PubMed]
- Feng J, Li J, Wu L, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res 2020;39:126. [Crossref] [PubMed]
- Wang XH, Jiang ZH, Yang HM, et al. Hypoxia-induced FOXO4/LDHA axis modulates gastric cancer cell glycolysis and progression. Clin Transl Med 2021;11:e279. [Crossref] [PubMed]
- Su X, Yang Y, Yang Q, et al. NOX4-derived ROS-induced overexpression of FOXM1 regulates aerobic glycolysis in glioblastoma. BMC Cancer 2021;21:1181. [Crossref] [PubMed]
- Raja E, Morikawa M, Nishida J, et al. Tyrosine kinase Eph receptor A6 sensitizes glioma-initiating cells towards bone morphogenetic protein-induced apoptosis. Cancer Sci 2019;110:3486-96. [Crossref] [PubMed]
- Zhang L, Zhang J, Ye Z, et al. Isoflavone ME-344 Disrupts Redox Homeostasis and Mitochondrial Function by Targeting Heme Oxygenase 1. Cancer Res 2019;79:4072-85. [Crossref] [PubMed]
- Chau LY. Heme oxygenase-1: emerging target of cancer therapy. J Biomed Sci 2015;22:22. [Crossref] [PubMed]
- Lou JS, Zhao LP, Huang ZH, et al. Ginkgetin derived from Ginkgo biloba leaves enhances the therapeutic effect of cisplatin via ferroptosis-mediated disruption of the Nrf2/HO-1 axis in EGFR wild-type non-small-cell lung cancer. Phytomedicine 2021;80:153370. [Crossref] [PubMed]
- Chen D, Wu YX, Qiu YB, et al. Hyperoside suppresses hypoxia-induced A549 survival and proliferation through ferrous accumulation via AMPK/HO-1 axis. Phytomedicine 2020;67:153138. [Crossref] [PubMed]
- Tertil M, Golda S, Skrzypek K, et al. Nrf2-heme oxygenase-1 axis in mucoepidermoid carcinoma of the lung: Antitumoral effects associated with down-regulation of matrix metalloproteinases. Free Radic Biol Med 2015;89:147-57. [Crossref] [PubMed]
- McKeen HD, Byrne C, Jithesh PV, et al. FKBPL regulates estrogen receptor signaling and determines response to endocrine therapy. Cancer Res 2010;70:1090-100. [Crossref] [PubMed]
- Donley C, McClelland K, McKeen HD, et al. Identification of RBCK1 as a novel regulator of FKBPL: implications for tumor growth and response to tamoxifen. Oncogene 2014;33:3441-50. [Crossref] [PubMed]
- Di X, Jin X, Ma H, et al. The Oncogene IARS2 Promotes Non-small Cell Lung Cancer Tumorigenesis by Activating the AKT/MTOR Pathway. Front Oncol 2019;9:393. [Crossref] [PubMed]
- Yin Y, Liu Y, Wang Y, et al. DZIP1 expressed in fibroblasts and tumor cells may affect immunosuppression and metastatic potential in gastric cancer. Int Immunopharmacol 2023;117:109886. [Crossref] [PubMed]
- Cui XB, Shen YY, Jin TT, et al. SLC39A6: a potential target for diagnosis and therapy of esophageal carcinoma. J Transl Med 2015;13:321. [Crossref] [PubMed]
- Cheng X, Wei L, Huang X, et al. Solute Carrier Family 39 Member 6 Gene Promotes Aggressiveness of Esophageal Carcinoma Cells by Increasing Intracellular Levels of Zinc, Activating Phosphatidylinositol 3-Kinase Signaling, and Up-regulating Genes That Regulate Metastasis. Gastroenterology 2017;152:1985-1997.e12. [Crossref] [PubMed]
- In 't Veld SGJG, Arkani M, Post E, et al. Detection and localization of early- and late-stage cancers using platelet RNA. Cancer Cell 2022;40:999-1009.e6. [Crossref] [PubMed]
- Zhang J, Xu R, Lu Q, et al. A Novel Methylation Marker NRN1 plus TERT and FGFR3 Mutation Using Urine Sediment Enables the Detection of Urothelial Bladder Carcinoma. Cancers (Basel) 2023;15:615. [Crossref] [PubMed]
- Heinhuis KM, In 't Veld SGJG, Dwarshuis G, et al. RNA-Sequencing of Tumor-Educated Platelets, a Novel Biomarker for Blood-Based Sarcoma Diagnostics. Cancers (Basel) 2020;12:1372.
- Kang J, Choi YJ, Kim IK, et al. LASSO-Based Machine Learning Algorithm for Prediction of Lymph Node Metastasis in T1 Colorectal Cancer. Cancer Res Treat 2021;53:773-83. [Crossref] [PubMed]
- Suarez-Cabrera C, Estudillo L, Ramón-Gil E, et al. BlaDimiR: A Urine-based miRNA Score for Accurate Bladder Cancer Diagnosis and Follow-up. Eur Urol 2022;82:663-7. [Crossref] [PubMed]
- Lefrançais E, Ortiz-Muñoz G, Caudrillier A, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 2017;544:105-9. [Crossref] [PubMed]
- Roweth HG, Battinelli EM. Lessons to learn from tumor-educated platelets. Blood 2021;137:3174-80. [Crossref] [PubMed]
- Risitano A, Beaulieu LM, Vitseva O, et al. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood 2012;119:6288-95. [Crossref] [PubMed]
- Wu X, Zhang B, Chen K, et al. Baffled-flow culture system enables the mass production of megakaryocytes from human embryonic stem cells by enhancing mitochondrial function. Cell Prolif 2023;56:e13484. [Crossref] [PubMed]
- Garcia A, Dunoyer-Geindre S, Nolli S, et al. miR-204-5p and Platelet Function Regulation: Insight into a Mechanism Mediated by CDC42 and GPIIbIIIa. Thromb Haemost 2021;121:1206-19. [Crossref] [PubMed]
- Xing L, Tang X, Wu K, et al. TRIM27 Functions as a Novel Oncogene in Non-Triple-Negative Breast Cancer by Blocking Cellular Senescence through p21 Ubiquitination. Mol Ther Nucleic Acids 2020;22:910-23. [Crossref] [PubMed]
- Comandatore A, Franczak M, Smolenski RT, et al. Lactate Dehydrogenase and its clinical significance in pancreatic and thoracic cancers. Semin Cancer Biol 2022;86:93-100. [Crossref] [PubMed]
- Sharma D, Singh M, Rani R. Role of LDH in tumor glycolysis: Regulation of LDHA by small molecules for cancer therapeutics. Semin Cancer Biol 2022;87:184-95. [Crossref] [PubMed]
- Hua Q, Jin M, Mi B, et al. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol 2019;12:91. [Crossref] [PubMed]
- Xie M, Fu XG, Jiang K. Notch1/TAZ axis promotes aerobic glycolysis and immune escape in lung cancer. Cell Death Dis 2021;12:832. [Crossref] [PubMed]
- Cao L, Wu J, Qu X, et al. Glycometabolic rearrangements--aerobic glycolysis in pancreatic cancer: causes, characteristics and clinical applications. J Exp Clin Cancer Res 2020;39:267. [Crossref] [PubMed]
- Cai K, Chen S, Zhu C, et al. FOXD1 facilitates pancreatic cancer cell proliferation, invasion, and metastasis by regulating GLUT1-mediated aerobic glycolysis. Cell Death Dis 2022;13:765. [Crossref] [PubMed]
- Khajah MA, Khushaish S, Luqmani YA. Lactate Dehydrogenase A or B Knockdown Reduces Lactate Production and Inhibits Breast Cancer Cell Motility in vitro. Front Pharmacol 2021;12:747001. [Crossref] [PubMed]
- Cascardo F, Anselmino N, Páez A, et al. HO-1 Modulates Aerobic Glycolysis through LDH in Prostate Cancer Cells. Antioxidants (Basel) 2021;10:966. [Crossref] [PubMed]
- Pajak B, Siwiak E, Sołtyka M, et al. 2-Deoxy-d-Glucose and Its Analogs: From Diagnostic to Therapeutic Agents. Int J Mol Sci 2019;21:234. [Crossref] [PubMed]


