
Role of CENPL, DARS2, and PAICS in determining the prognosis of patients with lung adenocarcinoma
Highlight box
Key findings
• This study showed that aspartyl-tRNA synthetase 2 (DARS2) and phosphoribosylaminoimidazole carboxylase (PAICS) expression were significantly increased in lung adenocarcinoma (LUAD) tissues. Expression of centromere protein L (CENPL), DARS2, and PAICS was negatively correlated with LUAD patients prognosis. We successfully established a prognostic model that integrated DARS2, PAICS, and other clinicopathological variables (age, gender, and tumor stage) to effectively predict the prognosis of LUAD patients.
What is known and what is new?
• LUAD is the most prevalent histological subtype of non-small cell lung cancer. In recent years, remarkable progress has been made in multimodal treatment strategies. However, the 5-year overall survival rate of advanced LUAD patients remains poor. To accurately estimate the individual survival of LUAD patients, many independent prognostic factors, including age, gender and histology, have been identified.
• This study successfully identified 30 differentially expressed genes that may be associated with the pathogenesis of LUAD. Among these genes, DARS2 and PAICS were identified as prognostic biomarkers. Furthermore, we comprehensively analyzed the expression and prognostic value of CENPL, DARS2, and PAICS. Finally, a prediction model was successfully constructed based on clinical characteristics combined with DARS2 and PAICS.
What is the implication, and what should change now?
• This study comprehensively analyzed the expression and prognostic value of CENPL, DARS2, and PAICS for LUAD. Based on these data, a robust and stable prognostic nomogram was successfully established by integrating DARS2, PAICS, and other clinicopathological variables (age, gender, and tumor stage). This promising model may serve as a reference for clinicians and help them to select more effective interventions.
Introduction
Non-small cell lung cancer (NSCLC), which accounts for about 85% of lung cancers, is the leading cause of tumor-related mortality worldwide (1,2). The proportions of NSCLC histological subtypes vary according to race. Among all NSCLC subtypes, Lung adenocarcinoma (LUAD) accounted for almost 47% of cases in Western patients, while about 55–60% of cases in Chinese patients (3). LUAD is the most prevalent histological subtype in NSCLC, and accounts for 40% of lung cancer cases (4,5). In recent years, significant progress has been made in multimodal treatment strategies, including surgical resection, chemotherapy, radiotherapy, immunotherapy, and molecular targeted therapy.
A study had verified that some independent prognostic factors, including age, gender and histology, can be used to partially predict individual survival of lung cancer patients (6). However, there are still many limitations. For instance, the predictive accuracy of a single independent prognostic factor is limited. Combining multiple independent prognostic factors could improve the accuracy of prediction. To enhance the accuracy of these estimates, Cox proportional hazards models have been widely adopted (7). For instance, a nomogram is a reliable tool that has the function of quantifying risk by combining and clarifying important clinical characteristics of patients (8). By drawing a concise graph of the outcome-risk predictive model, the nomogram derives the risk probability of a specific event, such as lung cancer-specific survival. Multiple studies have confirmed that nomograms have the ability to predict prognosis in various cancers (9-11). As a 5-year overall survival (OS) rate in advanced LAUD patients remains less than 15% (12), often due to local recurrence and distant metastasis, there is an urgent need to explore novel prognostic biomarkers and construct an effective prognostic model.
Therefore, we conducted a comprehensive bioinformatic analysis and aimed to explore promising biomarkers and construct a prognostic model for the LUAD. Finally, we successfully established a robust and stable prognostic nomogram by integrating DARS2, PAICS, and other clinicopathological variables (age, gender, and tumor stage). This promising model may serve as a reference for clinicians and help them to select more effective interventions. We present this article in accordance with the TRIPOD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-696/rc).
Methods
Ethics statements
This study was the research of public databases and was a retrospective study. The study protocol was approved by the Ethical Committee of the Affiliated Hospital of Qingdao University (No. QYFY WZLL 28933), and informed consent was obtained from all the participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was a single center study (The Affiliated Hospital of Qingdao University). Patients who underwent lobectomy or lung segmentectomy in our hospital and had pathological invasive adenocarcinoma were included in this study.
Data retrieving and processing
The Genomic Data Commons-The Cancer Genome Atlas-Lung Adenocarcinoma (GDC-TCGA-LUAD) dataset (Data Release 18.0, July 8, 2019) was downloaded from the University of California, Santa Cruz (UCSC) Xena browser. The data type count and Fragments Per Kilobase of transcript per Million mapped reads (FPKM) were selected to extract the “primary solid tumor” and convert it into transcripts per million (TPM) format. “Masked Somatic Mutation” data were selected as somatic mutation data of LUAD patients. VarScan software (v2.4.0) was used to pre-process the data, and maftools R package (13) was used to visualize the somatic mutation data. At the same time, clinical characteristics of LUAD patients, including age, TNM stage, survival time, and survival status information, were also downloaded from GDC-TCGA-LUAD dataset. Missing clinical information was excluded from the study.
GSE72094 (14) and GSE13213 (15) gene expression data and corresponding clinical information (including survival time and survival state) were downloaded from the Gene Expression Omnibus (GEO) database. Data samples of homo sapiens were used. The GSE72094 chip platform was based on GPL15048. Samples without survival information were removed. Ultimately, 398 surgical specimens were included in this study. The GSE13213 chip platform was based on GPL6480. Samples without survival information were discarded. Ultimately, 117 tumor samples were retained in this study. Limma R package (16) was used to standardize the data.
Identification of DEGs
First, cancer and normal samples were extracted from TCGA dataset. The DESeq2 R package (17) was used for differential analysis of count data, and a log fold change (FC) >1 and an adjusted (adj.) P<0.05 were set as cut-off values. Second, the cancer samples were further divided into early (stage I and II) and late (stage III and IV) stage groups. Again, DESeq2 R package was used for differential analysis, and a logFC >1 and an adj. P<0.05 were used as cut-off values. Third, the GSE72094 and GSE13213 datasets were divided into early and late stage groups based on tumor stage. Limma R package (16) was used for differential analysis, and a logFC >1 and an adj. P<0.05 were chosen as cut-off values. All the statistical P values were bilateral, and a P value <0.05 was considered statistically significant.
Gene set enrichment analysis and protein-protein interaction (PPI) network
We used the OmicShare Tools (www.omicshare.com/tools) to conduct Gene Ontology (GO) (18) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (19) enrichment analyses of obtained DEGs. In addition, to explore possible relationships among DEGs, a PPI network of DEGs was constructed through the STRING database (20).
Identification of key genes significantly associated with prognosis
To further assess the prognostic value of identified DEGs and identify reliable prognostic biomarkers, a survival analysis was performed using the TCGA-LUAD, GSE72094, and GSE13213 datasets. The median gene expression was set as cut-off value to compare differences in survival rates. The Kaplan-Meier method was used for survival analysis. And a P value <0.05 was considered statistically significant. Three groups of prognostic genes were intersected to identify key prognostic genes that may be closely related to development of LUAD and thus could serve as prognostic biomarkers. All the statistical P values were bilateral, and a P value <0.05 was considered statistically significant.
Multi-omics analysis of the key prognostic genes
A multi-omics analysis of key prognostic genes was performed using the Gene Set Cancer Analysis (GSCA, http://bioinfo.life.hust.edu.cn/GSCA). This analysis aimed to further investigate the copy number variations (CNVs), single nucleotide variations (SNVs), and methylation changes of key prognostic genes, and explore the prognostic differences and their relationship with transcriptomes. All the statistical P values were bilateral, and a P value <0.05 was considered statistically significant.
Relationship between key prognostic genes and immune infiltration
Using RNA-sequencing data from TCGA, the Tumor Immune Estimation Score (TIMER) (21) (https://cistrome.shinyapps.io/timer/) database was used to estimate the relationship between gene expression and immune cell level of tumor infiltration. TIMER was used to calculate the association between expression of key genes and tumor purity and immune cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophil, macrophages, and dendritic cells. All the statistical P values were bilateral, and a P value <0.05 was considered statistically significant.
Relationship between key prognostic genes and drug sensitivity
The data, including messenger RNA (mRNA) expression profiles of genes and pharmacological activity data, were downloaded from CellMiner database (https://discover.nci.nih.gov/cellminer/) (22). A Spearman correlation analysis was conducted to assess the correlation between expression of key genes and sensitivity of the chemical compounds. A P value <0.05 was considered statistically significant. Additionally, relevant data were obtained from the Genomics of Drug Sensitivity in Cancer (GDSC, https://www.cancerrxgene.org) and the Cancer Therapeutics Response Portal (CTRP, version 2, https://portals.broadinstitute.org/ctrp/) databases. From the GDSC database, we downloaded gene expression profiles of various tumor cell lines along with their corresponding half-maximal inhibitory concentration (IC50) values for specific drugs to assess drug sensitivity. From the CTRP database, we retrieved area under the curve (AUC) values representing cell viability following drug treatment. To investigate the relationship between key gene expression and drug sensitivity, we first matched the gene expression data with the drug sensitivity data to ensure consistency across cell lines. Subsequently, both gene expression levels and drug sensitivity metrics (IC50 or AUC) were normalized. Spearman’s rank correlation analysis was employed to assess the non-parametric correlation between key gene expression levels and drug sensitivity. All statistical analyses were conducted using R software (version 4.0.3). All tests were two-sided, and a P value less than 0.05 was considered statistically significant.
Construction and evaluation of the nomogram and prognostic model
To determine whether key genes were independent prognostic factors of LUAD, univariate and multivariate Cox regression was further performed to analyze key genes and other clinical characteristics (e.g., age, sex, and stage) of the TCGA-LUAD dataset. A P value <0.05 indicated that the factor was an independent prognostic factor of LUAD in multivariate Cox regression. Next, a nomogram was constructed based on optimal multivariate Cox regression analysis to predict 1-, 3-, and 5-year survival probabilities of LUAD patients. The rms R package (https://CRAN.R-project.org/package=rms) was used to construct the nomogram. The time-dependent receiver operating characteristic (ROC) and calibration curves were used to evaluate accuracy and consistency of the model. In addition, as the clinical characteristics of GSE72094 could not be matched with TCGA-LUAD, the GSE13213 dataset was used to test the model, and the time-dependent ROC curve was used to evaluate accuracy of the test set. All the statistical P values were bilateral, and a P value <0.05 was considered statistically significant.
Immunohistochemistry
Samples were obtained from LUAD patients. Both paired tumor and adjacent normal lung tissues were collected from each patient. All samples were resected and immediately placed in liquid nitrogen. Four-µm-thick sections were deparaffinized and rehydrated, and endogenous peroxidases were blocked with 3% hydrogen peroxide at room temperature. The sections were then incubated with 3% bovine serum albumin (GC305010, Wuhan Servicebio Technology Co., Ltd., Wuhan, China). Rabbit polyclonal anti-PAICS antibody (12967-1-ap, Proteintech) and rabbit polyclonal anti-DARS2 (13807-1-ap, Proteintech) were each incubated in a 1:200 dilution of phosphate-buffered saline (PBS) at 4 °C overnight, and then incubated with a horseradish peroxidase (HRP)-coupled anti-rabbit polymer, after which diaminobenzidine detection was performed (G1212, Wuhan Servicebio Technology Co., Ltd.). The sections were counterstained with hematoxylin. Both PAICS and DARS2 immunoreactivity in the tumor and adjacent normal lung tissues was evaluated with light microscope. At last, the differences of PAICS and DARS2 protein expression between normal lung tissues and LUAD specimens were quantified and significantly analyzed
Statistical analysis
The statistical analysis was performed using R software (https://www.r-project.org/, version 4.1.1). For comparisons of two groups of continuous variables, the independent Student’s t-test was used to estimate statistical significance of the normally distributed variables, and the Mann-Whitney U test (Wilcoxon rank-sum test) was used to analyze differences between non-normally distributed variables. All the statistical P values were bilateral, and a P value <0.05 was considered statistically significant.
Results
Patient characteristics
In this study, the characteristics of LUAD patients were shown in Table 1. Two hundred and seventy-eight patients diagnosed with stage I LUAD were identified. One hundred and twenty-four patients diagnosed with stage II LUAD were identified. Eighty-three patients diagnosed with stage III LUAD were identified. Twenty-seven patients diagnosed with stage IV LUAD were identified.
Table 1
| Characteristics | Values |
|---|---|
| Stage† | |
| IV | 27 (5.3) |
| I | 278 (54.3) |
| III | 83 (16.2) |
| II | 124 (24.2) |
| OS | |
| Living | 328 (63.8) |
| Deceased | 186 (36.2) |
| Overall survival (months) | 21.468 (13.644, 37.15) |
| Sex | |
| Male | 239 (46.5) |
| Female | 275 (53.5) |
Values are presented as n (%) or median (IQR). †, 2 patients in the database did not have a TNM stage. LUAD, lung adenocarcinoma; OS, overall survival; IQR, interquartile range.
Identification of DEGs
First, the count data of TCGA-LUAD were standardized using the DESeq2 package. After standardization, a principal component analysis (PCA) of normal and tumor samples was conducted and results showed that there was a significant difference between both groups (Figure 1A). The PCA also showed that there was no discrimination between early and late stage groups (Figure 1B). Analyzing DEGs of normal and tumor samples of TCGA-LUAD, as well as early and late stage groups of TCGA-LUAD, GSE13213, and GSE72094, identified the following 30 DEGs: OTUD1, GINS1, PAICS, GPD1, PPP1R15A, SIRPB1, TRPV2, AGRP, CENPL, PRAM1, DARS2, GART, ILF2, NCF2, ALOX5AP, CYP27A1, NELL1, CD300LF, HK3, RTN1, MNDA, CNTNAP2, PLLP, TUBA4A, CD37, ENHO, EVX1, FAM180A, FHAD1, and NME9 (Figure 1C). We used heat maps to display 30 DEGs in TCGA-LUAD, GSE72094, and GSE13213 datasets (Figure 1D-1F).

Functional enrichment analysis and PPI network construction
The GO results showed that DEGs were mainly enriched in BPs and MFs (Figure 2A). The top three GO enrichment results were as follows: GO0004637: phosphoribosylamine-glycine ligase activity; GO0004638: PAICS activity; and GO0004639: phosphoribosylaminoimidazole succinocarboxamide synthase activity. KEGG analysis showed that the top three enriched pathways were: ko00524: neomycin, kanamycin and gentamicin biosynthesis, ko04380: osteoclast differentiation; and ko00230: purine metabolism (Figure 2B).

In the PPI network, a total of 30 nodes were connected. There were also a total of 9 edges, and the average node rate was 0.6. The P value of the PPI enrichment analysis was 0.01 (Figure 2C).
Identification of key genes significantly associated with prognosis
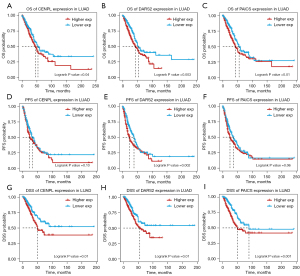
The results of prognostic analysis revealed common prognostic genes in the three datasets: CENPL, DARS2 and PAICS (Figure 3A). Expression levels of CENPL, DARS2, and PAICS were negatively correlated with prognosis of LUAD patients in the GSE13213 and GSE72094 datasets (Figure 3B-3G). In TCGA-LUAD dataset, OS, progression-free survival (PFS) and disease-specific survival (DSS) were selected for survival analysis. In the survival analysis of OS and DSS, a high expression of CENPL, DARS2, and PAICS indicated a poorer prognosis. However, in the survival analysis of PFS, only a high expression of DARS2 indicated a poorer prognosis. (Figure 4A-4I).

Multi-omics analysis of key prognostic genes
To further understand the characteristics and potential mechanisms of these key prognostic genes in the pathogenesis of LUAD, a multi-omics analysis was performed. In pan-cancer expression profiles, expression levels of CENPL and PAICS were increased in all tumor tissues, except for thyroid cancer tissues. Expression of DARS2 was also significantly up-regulated in all tumor tissues, except for thyroid cancer, renal clear cell carcinoma, and renal papillary carcinoma tissues. Expression of CENPL, DARS2, and PAICS was significantly higher in tumor tissues of LUAD and lung squamous cell carcinoma compared to normal lung tissues (Figure 5A-5C). Furthermore, CENPL, DARS2, and PAICS were all highly expressed in late stage groups (Figure 5D-5F) compared with early stage, especially in patients with stage IV disease (Figure 5G).

To comprehensively analyze their characteristics of CENPL, DARS2 and PAICS, changes in their genome levels were analyzed and revealed that the mutation sites of all three genes were rare and missense mutations (Figure 6A-6C).

CNV analysis showed that a greater proportion of copy number amplification (including heterozygous and pure sum amplification) was observed in CENPL and DARS2 compared to PAICS, while the proportion of copy number amplification and deletion was similar in PAICS (Figure 6D-6F). A correlation analysis between CNV and mRNA expression levels of the transcriptome data showed that CNV of DARS2 was significantly positively correlated with its mRNA expression (Figure 6G-6I). However, CNV prognostic analysis of CENPL, DARS2, and PAICS showed that there was no significant prognostic difference between amplified, deleted, and wild type mutations (Table S1). In addition, significant differences were observed in methylation of all three genes between LUAD and normal lung tissues. However, there was no significant difference in all three genes’ prognosis (Tables S2,S3).
The relationship between key prognostic genes and immune infiltration
Expression levels of CENPL, DARS2, and PAICS were not significantly correlated with tumor purity, B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophil, or dendritic cells (Figure 7A-7C). Subsequently, when analyzing the relationship between CNVs of CENPL, DARS2, and PAICS, and immune cell infiltration, we found that immune cell infiltration tended to be decreased in CNV amplified types (Figure S1A-S1C).

Relationship between key prognostic genes and drug sensitivity
As the CENPL, DARS2, and PAICS genes were elevated in the late stage LUAD patients, especially in stage IV patients, but not associated with immune cells, we combined drug and small molecule compound data from the GDSC and CTRP datasets. Drug sensitivity of CENPL, DARS2, and PAICS in the GCP, GDSC, and CTRP datasets was analyzed by cellMiner. In the GCP dataset, we found that CENPL was positively correlated with calusterone, nelarabine, fenretinide, and rapamycin, DARS2 was positively correlated with vorinostat, fulvestrant, parthenolide, and allopurinol, and PAICS was positively correlated with chelerythrine, amonafide, pyrazoloacridine, and nelarabine (Figure 8A-8C). In the GDSC dataset, we found that CENPL and PAICS were positively correlated with 17-AAG, afatinib, and gefitinib, while CENPL and PAICS were negatively correlated with BIX02189, BMS345541, and BX-912 (Figure 8D). In the CTRP dataset, Results showed that the expression of CENPL, DARS2, and PAICS was negatively correlated with the response of most drugs, including BI-2536, CD-437, and CHM-1 (Figure 8E).

Construction and evaluation of the nomogram and prognostic model
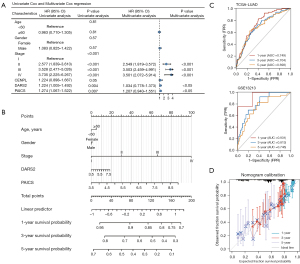
The univariable Cox analysis showed that DARS2, PAICS and tumor stage were determined to be independent prognostic factors of LUAD (P<0.01). On the contrary, CENPL was not a prognostic factor. These results were confirmed in the multivariable Cox analysis (P<0.05) (Figure 9A). Second, a prognostic nomogram was established integrating age, gender, DARS2, PAICS, and tumor stage (Figure 9B). The 1-, 3-, and 5-year areas under the curve (AUCs) of the training set were 0.749, 0.704, and 0.698, respectively. The 1-, 3-, and 5-year AUCs of the test set were 0.931, 0.813, and 0.746, respectively (Figure 9C). In addition, calibration curves of the 1-, 3-, and 5-year survival rates showed an optimal agreement between nomogram prediction and actual observations (Figure 9D).
Immunohistochemistry
Immunohistochemistry analysis of LUAD histological samples showed a higher expression of DARS2 and PAICS in malignant samples compared with normal lung tissue (Figure 10) (the independent Student’s t-test, P<0.05).

Discussion
In the past decades, major breakthroughs have been made in LUAD treatment. However, the long-term clinical outcomes of advanced LUAD patients have remained poorer than expected. Due to delayed diagnosis, high recurrence rate, and drug resistance, the 5-year OS rate of advanced LUAD patients remains less than 15% (12). This undermines an unmet need for effective prognostic biomarkers.
In the present study, we comprehensively analyzed the expression and prognostic value of CENPL, DARS2, and PAICS in LUAD patients in multiple databases using bioinformatic analyses. These results suggested that CENPL, DARS2, and PAICS might be oncogenes, and their genes are amplified to increase the host gene expression and promote tumor progression and metastasis. In addition, we successfully constructed a prediction model for OS based on LUAD clinical characteristics, DARS2, and PAICS. This model is promising and may serve as a reference for clinicians and help them to make better clinical decisions when treating LUAD patients.
CENPL is a member of the centromere protein (CENP) family and plays important roles in regulating cell division. CENPs are crucial members of the centromere and kinetochore, which determine the separation of chromosomes during mitosis and meiosis (23). A previous study confirmed that several CENPs are highly expressed in LUAD tissues compared to normal lung tissues and have significant prognostic value (24).
DARS2, which is encoded by the class-II aminoacyl-tRNA synthetase family gene, is a mitochondrial enzyme that specifically aminoacylates aspartyl-tRNA (25). DARS2 is located in chromosome 1q25.1, which mainly exists in the liver, spinal cord and brain stem (26-28). A previous study showed that the expression of DARS2 was significantly upregulated in LUAD tissues, and DARS2 plays a role in LUAD by targeting the ERK/c-Myc signaling pathway (29).
PAICS is a de novo purine metabolic enzyme that generates N-succinocarboxyamide-5-aminoimidazole ribonucleotide via using 5-aminoimidazole ribonucleotide (AIR) by adenylosuccinate lyase. Previous research has shown that PAICS is significantly overexpressed in LUAD tissues and is a putative prognostic biomarker of LUAD (30). In addition, PAICS has also been shown to play an oncogenic role in EGFR wild-type NSCLC and represent a potential therapeutic target (31).
In recent years, there has been remarkable progress in treatment modes for lung cancer. Currently, molecular targeted therapy and immunotherapy are receiving more and more attention. Immunotherapy is rapidly gaining popularity in the armamentarium of treatment modalities for lung cancer. The most recent and very promising development is the use of neoadjuvant chemo-immunotherapy in resectable NSCLC (32). Our results suggested that all three genes may not be associated with the activation or suppression of immune cells. However, we have successfully discovered a variety of sensitive drugs associated with CENPL, DARS2, and PAICS. The mechanism of these genes needs to be further studied in the future.
In this study, we found CENPL, DARS2, and PAICS are predictive biomarkers in LUAD patients. This study did not only analyzed the molecular differences between early and late stage LUAD at the transcriptome level, but also explored the differential expression of CENPL, DARS2, and PAICS, and related differences in tumor microenvironment by conducting both genomics and immune infiltration analyses. The combined application of multi-omics and multi-analysis methods could greatly improve the accuracy of predicting marker molecules. However, this study also had some limitations. The data in this study were derived from public databases and multiple datasets were jointly used to improve the accuracy of the results. Although we have performed an in-depth analysis based on the available data set, it is undeniable that the limitation of the database may affect the accuracy of the results. Therefore, we expect that more real-world studies or prospective clinical trials could improve the above data and conduct a more comprehensive analysis to further verify the results of this study. To properly translate these biomarkers into clinically usable molecular markers, it is necessary to expand the sample and adopt prospective experimental methods to validate our findings.
Conclusions
In conclusion, we successfully identified 30 DEGs that may be associated with the pathogenesis of LUAD. Among these genes, DARS2 and PAICS were identified as biomarkers that could predict the LUAD prognosis. We comprehensively analyzed the expression and prognostic value of CENPL, DARS2, and PAICS for LUAD. Additionally, a prediction model was successfully constructed based on clinical characteristics combined with DARS2 and PAICS.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-696/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-696/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-696/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-696/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethical Committee of the Affiliated Hospital of Qingdao University (No. QYFY WZLL 28933), and informed consent was obtained from all the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Li Y, Yu X, Zhang Y, et al. Identification of a novel prognosis-associated ceRNA network in lung adenocarcinoma via bioinformatics analysis. Biomed Eng Online 2021;20:117. [Crossref] [PubMed]
- Chen P, Liu Y, Wen Y, et al. Non-small cell lung cancer in China. Cancer Commun (Lond) 2022;42:937-70. [Crossref] [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Shi J, Hua X, Zhu B, et al. Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study. PLoS Med 2016;13:e1002162. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Xie D, Marks R, Zhang M, et al. Nomograms Predict Overall Survival for Patients with Small-Cell Lung Cancer Incorporating Pretreatment Peripheral Blood Markers. J Thorac Oncol 2015;10:1213-20. [Crossref] [PubMed]
- She Y, Jin Z, Wu J, et al. Development and Validation of a Deep Learning Model for Non-Small Cell Lung Cancer Survival. JAMA Netw Open 2020;3:e205842. [Crossref] [PubMed]
- Wu J, Zhang H, Li L, et al. A nomogram for predicting overall survival in patients with low-grade endometrial stromal sarcoma: A population-based analysis. Cancer Commun (Lond) 2020;40:301-12. [Crossref] [PubMed]
- Lv J, Liu YY, Jia YT, et al. A nomogram model for predicting prognosis of obstructive colorectal cancer. World J Surg Oncol 2021;19:337. [Crossref] [PubMed]
- Li Y, Chen D, Xuan H, et al. Construction and validation of prognostic nomogram for metaplastic breast cancer. Bosn J Basic Med Sci 2022;22:131-9. [Crossref] [PubMed]
- Yang Z, Liu B, Lin T, et al. Multiomics analysis on DNA methylation and the expression of both messenger RNA and microRNA in lung adenocarcinoma. J Cell Physiol 2019;234:7579-86. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209-16. [Crossref] [PubMed]
- Tomida S, Takeuchi T, Shimada Y, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol 2009;27:2793-9. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Gene Ontology Consortium. going forward. Nucleic Acids Res 2015;43:D1049-56. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Luna A, Elloumi F, Varma S, et al. CellMiner Cross-Database (CellMinerCDB) version 1.2: Exploration of patient-derived cancer cell line pharmacogenomics. Nucleic Acids Res 2021;49:D1083-93. [Crossref] [PubMed]
- Feng Z, Chen Y, Cai C, et al. Pan-Cancer and Single-Cell Analysis Reveals CENPL as a Cancer Prognosis and Immune Infiltration-Related Biomarker. Front Immunol 2022;13:916594. [Crossref] [PubMed]
- Wang Y, Chen J, Meng W, et al. A five-gene expression signature of centromeric proteins with prognostic value in lung adenocarcinoma. Transl Cancer Res 2023;12:273-86. [Crossref] [PubMed]
- van Berge L, Hamilton EM, Linnankivi T, et al. Leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation: clinical and genetic characterization and target for therapy. Brain 2014;137:1019-29. [Crossref] [PubMed]
- Yahia A, Elsayed L, Babai A, et al. Intra-familial phenotypic heterogeneity in a Sudanese family with DARS2-related leukoencephalopathy, brainstem and spinal cord involvement and lactate elevation: a case report. BMC Neurol 2018;18:175. [Crossref] [PubMed]
- N'Gbo N'Gbo Ikazabo R, Mostosi C, Jissendi P, et al. A New DARS2 Mutation Discovered in an Adult Patient. Case Rep Neurol 2020;12:107-13. [Crossref] [PubMed]
- Inoue Y, Tanaka H, Kasho K, et al. Chromosomal location of the DnaA-reactivating sequence DARS2 is important to regulate timely initiation of DNA replication in Escherichia coli. Genes Cells 2016;21:1015-23. [Crossref] [PubMed]
- Fang T, Jiang J, Yu W, et al. DARS2 promotes the occurrence of lung adenocarcinoma via the ERK/c-Myc signaling pathway. Thorac Cancer 2023;14:3511-21. [Crossref] [PubMed]
- Zhou S, Yan Y, Chen X, et al. Roles of highly expressed PAICS in lung adenocarcinoma. Gene 2019;692:1-8. [Crossref] [PubMed]
- Li Y, Zhu L, Mao J, et al. Genome-scale CRISPR-Cas9 screen identifies PAICS as a therapeutic target for EGFR wild-type non-small cell lung cancer. MedComm (2020) 2024;5:e483.
- Franzi S, Mattioni G, Rijavec E, et al. Neoadjuvant Chemo-Immunotherapy for Locally Advanced Non-Small-Cell Lung Cancer: A Review of the Literature. J Clin Med 2022;11:2629. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


