
Mutational differences between primary cancer tissue and circulating tumor cells in early-stage non-small cell lung cancer
Highlight box
Key findings
• The mutational profiles of primary cancer tissue and circulating tumor cells in resectable early-stage non-small cell lung cancer (NSCLC) were significantly different. The concordance rate between the two samples was 17.8% using 405-gene targeted panel sequencing.
What is known and what is new?
• Circulating tumor cells are known to exhibit distinct mutations from those of primary cancer tissue based on whole exome sequencing or single cell analysis.
• Our study performed targeted panel sequencing for 405 cancer-associated genes in matched samples of primary cancer tissue and circulating tumor cells from 45 patients with early-stage NSCLC. The mutational roles of circulating tumor cells were different from those of primary cancer tissue. The number of mutations in circulating tumor cells did not show statistically significant associations with stage or histology.
What is the implication, and what should change now?
• Mutations in circulating tumor cells occurred independently from those in primary cancer tissue; therefore, some mutations in circulating tumor cells might be associated with the epithelial-mesenchymal transition. Further research is needed on the relationship between circulating tumor cells and distant recurrence.
Introduction
Non-small cell lung cancer (NSCLC) is recognized as a malignant disease with a high recurrence rate, even with appropriate management (1). Circulating tumor cells (CTCs), as a kind of liquid biopsies, offer advantages such as reduced invasiveness and the ease of repeated sampling. These benefits position CTCs as a breakthrough in various areas, including prognosis prediction, treatment monitoring, and the detection of drug resistance, especially for advanced-stage disease (2,3). However, limited data are available regarding the potential diagnostic, predictive, and staging value of CTCs in early-stage NSCLC.
Distant recurrence of early-stage NSCLC frequently occurs despite curative surgery (4,5). CTCs are believed to directly relate to metastasis through the process of epithelial-mesenchymal transition (EMT) (6). Several studies have suggested that the presence of CTCs, as detected by immunofluorescence imaging, is associated with poor prognosis or recurrence in patients with NSCLC (7-10).
We thought that CTCs harbor unique gene mutations that promote the EMT of primary cancer tissue. Examining the mutational profiles of CTCs could reveal additional characteristics associated with distant recurrence in early-stage NSCLC. However, most studies have been limited to the sequencing analysis of cell-free DNA, immunofluorescence imaging of CTCs, or single-cell analysis of CTC (7,11-14). The rarity of CTCs has led to a paucity of research on their mutational profiles or on direct comparisons using targeted panel sequencing between CTCs and primary cancer tissue, particularly in the context of early-stage NSCLC.
Thus, this study was conducted to analyze DNA from CTCs and compare the findings with those of primary cancer tissue using 405-gene targeted panel sequencing in patients with resectable early-stage NSCLC. We present this article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-709/rc).
Methods
Ethical statement
The study protocol was approved by the Institutional Review Board of Seoul National University Bundang Hospital (approval No. B-2211-791-304) and conformed to the principles of the Declaration of Helsinki (as revised in 2013). All participants provided written informed consent before enrollment.
Patients
A total of 45 patients were enrolled who were postoperatively diagnosed with NSCLC at Seoul National University Bundang Hospital between September and December 2023. The following exclusion criteria were applied: (I) patients with a histologic type other than adenocarcinoma or squamous cell carcinoma; (II) patients diagnosed with another malignancy within the past 5 years; (III) patients suspected of having any distant metastasis; and (IV) patients who refused to participate in this study.
Specimen processing
Following the induction of anesthesia, a total of 10 mL of fresh blood was drawn from the peripheral arterial blood into an ACD-A tube for CTC analysis. After the surgical specimen was removed, primary tissue samples from both normal and cancerous areas, each measuring 5 mm × 5 mm × 5 mm, were excised. These samples were promptly transported to BeyondDx Inc. (Gwangmyeong, Republic of Korea) for CTC extraction, which was performed within 6 hours of collection. Subsequently, DNA from the CTCs and primary tissue from the normal and cancerous lung regions were sent to Geninus (Seoul, Republic of Korea) for targeted panel sequencing.
Primary cancer tissue sequencing
DNA was extracted from the normal and cancerous tissues using a QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA, USA). The concentration and purity of the DNA were assessed with a NanoDrop One UV-Vis spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Qubit 4.0 Fluorometer (Life Technologies, Grand Island, NY, USA). The degree of DNA degradation was evaluated using a 4200 TapeStation instrument (Agilent Technologies, Santa Clara, CA, USA). Subsequently, the DNA was used to construct a library with the Complete Library Preparation Kit (Twist Bioscience, San Francisco, CA, USA) and CancerSCAN panel probes (Geninus), targeting 405 cancer-associated genes (Box S1). Sequencing was performed with the NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Mutations in the primary cancer tissue were identified using the sequencing results from the normal lung tissue as controls.
CTC staining and sequencing
Peripheral blood mononuclear cells (PBMCs) were isolated from the ACD-A tube, which was stored at room temperature, using Ficoll-Hypaque density gradient centrifugation. For CTC staining analysis, CD45+ PBMCs were depleted using Dynabeads (Invitrogen, Waltham, MA, USA). The remaining cells were stained with anti-epithelial cell adhesion molecule (EpCAM)‑fluorescein isothiocyanate (FITC), anti‑cytokeratin (CK)‑FITC, and anti‑CD45‑phycoerythrin antibodies and were counterstained with 4',6-diamidino-2-phenylindole (DAPI). MCF7 and Jurkat cells served as staining controls for CTCs and PBMCs, respectively. Images were captured with a Cytation 5 imaging system (Biotek Inc., Winooski, VT, USA) and analyzed using Gen5 software to enumerate the CTCs. CTCs were identified as DAPI+/CD45− and either double-positive (EpCAM+/CK+) or single-positive (EpCAM+/CK− or EpCAM−/CK+). In contrast, PBMCs were identified as DAPI+, EpCAM−/CK−/CD45+ (Figure 1). Separately, candidate CTCs for sequencing were isolated using EpCAM, EGFR, and HER2 biomarkers for targeted capture. Genomic DNA from CTCs expressing any of these markers—EpCAM, EGFR, or HER2—was extracted and amplified with the REPLI-g Single Cell kit (Qiagen Inc.) according to the manufacturer’s protocol. The amplified DNA was used to construct a library, which was then sequenced using the CancerSCAN panel, following the same methodology applied to primary cancer tissue samples.
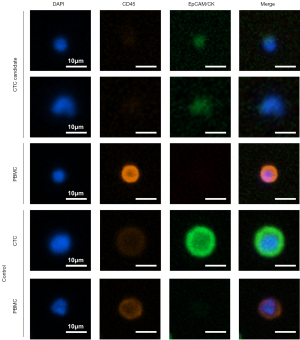
CTC positivity and mutation classification
On an immunofluorescent image, CTC positivity was defined as the presence of one or more CTCs per 2×106 PBMCs. For primary cancer tissue and CTCs, positive mutation status was indicated by the presence of one or more single nucleotide variant or insertion/deletion mutations. If a case exhibited at least one mutation common to primary cancer tissue and CTCs, it was considered to show partial concordance. Mutated genes were categorized based on their function; oncogenes included growth factor receptors, transcription factors, and signal transducers, while tumor suppressors consisted of genes functioning as caretakers and gatekeepers. A subset of genes, termed cancer hallmark genes, was identified using the methodology described by Liang et al. (15). Cancer hallmark genes were further classified into 11 categories or pathways: deregulated metabolism, evasion of anti-growth signaling, angiogenesis, immune system evasion, resistance to cell death, replicative immortality, sustained proliferative signaling, tissue invasion and metastasis, tumor-promoting inflammation, the tumor microenvironment, and genome instability. Comparative analysis was conducted using all available primary cancer tissue and CTC samples.
Statistical analysis
The primary objective of this study was to compare mutations between primary cancer tissue and CTCs based on sequencing profiles in early-stage NSCLC. The secondary objective was to examine the classification and roles of mutations between these samples. Categorical variables were reported as numbers and percentages, while continuous variables were expressed as medians with interquartile ranges (IQRs) and compared using the Student’s t-test. Discrete variables were analyzed using the chi-square method. Statistical analyses were performed using IBM SPSS Statistics for Windows, version 25 (IBM Corp., Armonk, NY, USA).
Results
Baseline and pathologic characteristics
The mean age of the 45 patients was 68.7±8.4 years, and 26 (57.8%) were male. Additional baseline and preoperative characteristics are detailed in Table S1. Of the patients, 37 (82.2%) were diagnosed with adenocarcinoma and 8 (17.8%) with squamous cell carcinoma. According to the American Joint Committee on Cancer, eighth edition, the pathologic stages were as follows: stage I in 30 patients (66.7%), stage II in 6 patients (13.3%), and stage III in nine patients (20.0%), as shown in Table 1.
Table 1
| Characteristics | Values (n=45), n (%) |
|---|---|
| Histology | |
| Adenocarcinoma | 37 (82.2) |
| Squamous cell carcinoma | 8 (17.8) |
| Pathologic stage | |
| I | 30 (66.7) |
| II | 6 (13.3) |
| III | 9 (20.0) |
| Lymph node metastasis | 9 (20.0) |
| Visceral pleural invasion | 15 (33.3) |
| Parietal pleural invasion | 3 (6.7) |
| Vascular invasion | 5 (11.1) |
| Lymphatic invasion | 11 (24.4) |
| Perineural invasion | 0 |
| Tumor spread through air spaces | 25 (55.6) |
| Necrosis | 18 (40.0) |
Panel sequencing data
Positive CTCs were identified by immunofluorescence imaging in 11 patients (24.4%), with a median count of 4.0 CTCs (IQR, 1.5–7.5). No clinical or pathologic factors were found to be associated with CTC positivity. Mutations were detected in all 45 primary cancer tissue samples (100%) and in 31 CTC samples (68.9%) (Figure 2). Regarding these mutations, a partial concordance rate of 17.8% was found between matched primary cancer tissue and CTCs.
Figure 3 presents the top 10 mutated genes in primary cancer tissues and CTCs. The most frequently mutated gene in primary cancer tissue was EGFR (n=17, 37.8%), followed by TP53 (n=12, 26.7%). In contrast, MSH6 mutations were found in all CTCs (n=31, 100%), with FANCE mutations being the second most prevalent among these cells (n=28, 90.3%). The mutation profiles of these top 10 genes in CTCs differed significantly from those in primary cancer tissues, with higher percentages for the mutations within CTCs.
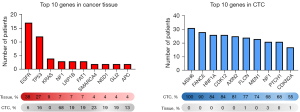
Table 2 presents the top 10 genes for primary cancer tissue and CTCs, categorized by pathologic stage and histologic type. In stage 1 disease, the most frequently mutated genes in primary cancer tissue were EGFR (n=14, 46.7%) and TP53 (n=6, 20.0%). In contrast, in stage 2 or higher, TP53 (n=6, 40.0%) and KRAS (n=4, 26.7%) were the most commonly mutated genes identified through panel sequencing. When primary cancer tissue was divided by histologic type, EGFR (n=17, 45.9%) and TP53 (n=8, 21.6%) were the predominant mutated genes in adenocarcinoma, while TP53 (n=4, 50.0%) and NF1 (n=3, 37.5%) were the most frequent in squamous cell carcinoma. For CTCs, MSH6 was the most commonly mutated gene, followed by FANCE, irrespective of stage or histologic type. The number of mutations in primary cancer tissue was higher in patients with a pathologic stage of 2 or above relative to stage 1 (P=0.008). However, the number of CTCs detected on immunofluorescent imaging and the mutation count in CTCs did not exhibit statistically significant differences (P=0.84 and P=0.56, respectively, Table 3). Additionally, no significant difference in the number of mutations was found in either sample across histologic types (Table 4).
Table 2
| Rank | Stage | Histology | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Stage 1 (n=30) | Stage >1 (n=15) | ADC (n=37) | SqCC (n=8) | ||||||||
| Genes | Number | Genes | Number | Genes | Number | Genes | Number | ||||
| Primary cancer tissue | |||||||||||
| 1 | EGFR | 14 | TP53 | 6 | EGFR | 17 | TP53 | 4 | |||
| 2 | TP53 | 6 | KRAS | 4 | TP53 | 8 | NF1 | 3 | |||
| 3 | NCOR1 | 2 | LRP1B | 3 | KRAS | 4 | NSD1 | 2 | |||
| 4 | NF1 | 2 | EGFR | 3 | LRP1B | 2 | NOTCH3 | 2 | |||
| 5 | BRAF | 2 | RB1 | 2 | RB1 | 2 | RB1 | 1 | |||
| 6 | KMT2D | 1 | PTPRD | 2 | PTPRD | 2 | PIK3CA | 1 | |||
| 7 | NSD1 | 1 | NOTCH2 | 2 | FAT1 | 2 | LRP1B | 1 | |||
| 8 | RB1 | 1 | SMARCA4 | 2 | GLI2 | 2 | KMT2D | 1 | |||
| 9 | PIK3CA | 1 | FAT1 | 2 | SMARCA4 | 2 | CHEK2 | 1 | |||
| 10 | NOTCH3 | 1 | PIK3CA | 1 | APC | 2 | FBXW7 | 1 | |||
| 11 | NTRK3 | 1 | NF1 | 1 | CREBBP | 2 | PAK5 | 1 | |||
| Circulating tumor cells | |||||||||||
| 1 | MSH6 | 18 | MSH6 | 13 | MSH6 | 26 | MSH6 | 5 | |||
| 2 | FANCE | 16 | FANCE | 12 | FANCE | 23 | FANCE | 5 | |||
| 3 | CDK12 | 16 | HNF1A | 12 | CDK12 | 22 | HNF1A | 5 | |||
| 4 | AXIN2 | 15 | FLCN | 11 | HNF1A | 21 | FLCN | 5 | |||
| 5 | HNF1A | 14 | CDK12 | 10 | AXIN2 | 21 | CDK12 | 4 | |||
| 6 | FLCN | 13 | AXIN2 | 10 | FLCN | 19 | AXIN2 | 4 | |||
| 7 | MEN1 | 13 | MEN1 | 10 | MEN1 | 19 | MEN1 | 4 | |||
| 8 | NF1 | 13 | NF1 | 8 | NF1 | 19 | CDKN2A | 3 | |||
| 9 | PTCH1 | 13 | PTCH1 | 8 | PTCH1 | 19 | NF1 | 2 | |||
| 10 | CDKN2A | 11 | CDKN2A | 6 | CDKN2A | 14 | PTCH1 | 2 | |||
| 11 | TERT | 6 | TERT | 5 | TERT | 9 | TERT | 2 | |||
ADC, adenocarcinoma; SqCC, squamous cell carcinoma.
Table 3
| Sample | Stage 1 (n=30) | Stage >1 (n=15) | P value |
|---|---|---|---|
| Immunofluorescent imaging of CTCs† | 5.0 [1.0–8.5] | 4.0 [3.0–4.0] | 0.84 |
| Mutations of primary cancer tissue | 6.0 [4.0–8.0] | 11.0 [7.0–14.0] | 0.008 |
| Mutations of CTCs | 27.0 [20.0–42.0] | 28.0 [22.0–43.5] | 0.56 |
Values are presented as median [interquartile range]. †, the count of CTCs was calculated only for patients (n=11) in whom these cells were detected by immunofluorescent imaging. CTCs, circulating tumor cells.
Table 4
| Sample | ADC (n=37) | SqCC (n=8) | P value |
|---|---|---|---|
| Immunofluorescent imaging of CTCs† | 4.0 [1.0–8.0] | 0.0 [0.0–0.0] | – |
| Mutations of primary cancer tissue | 7.0 [5.0–8.0] | 11.0 [8.5–15.0] | 0.17 |
| Mutations of CTCs | 28.5 [21.5–45.0] | 23.0 [20.5–36.5] | 0.45 |
Values are presented as median [interquartile range]. †, the count of CTCs was calculated only for patients (n=11) in whom these cells were detected by immunofluorescent imaging. CTCs, circulating tumor cells; ADC, adenocarcinoma; SqCC, squamous cell carcinoma.
Roles of mutated genes
Figure 4 presents the confirmed mutational profiles of genes in primary cancer tissue and CTCs, reflecting the roles of each gene. Primary cancer tissue exhibited mutations in both oncogenes, like growth factor receptors, and tumor suppressor genes, such as gatekeepers. In contrast, CTCs predominantly displayed mutations in tumor suppressor genes, including caretakers and gatekeepers, rather than oncogenes.
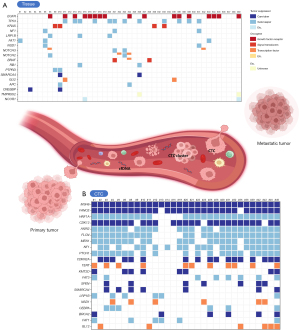
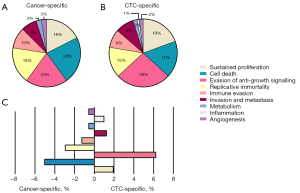
The hallmark pathways of cancer for each sample are depicted in Figure 5A,5B, while the proportional differences between the two samples are illustrated in Figure 5C. The hallmark pathways of the primary cancer tissue were characterized by sustained proliferation, resistance to cell death, and altered metabolism. In comparison, CTCs exhibited additional hallmark pathways, such as evasion of anti-growth signaling, replicative immortality, and invasion/metastasis.
Discussion
In this study, the concordance rate of gene mutation profiles between CTCs and the corresponding primary cancer tissue was relatively low in patients with early-stage NSCLC. The classifications and roles of the mutated genes were markedly different between the two sample types. These findings suggest that the mutational forces acting on CTC genomes may differ from those affecting primary cancer tissues. Given the scarcity of studies directly comparing primary cancer tissue and CTCs over 50 matched cases using panel sequencing, our study is meaningful. It represents an analysis of mutations in CTCs and primary cancer tissues, with panel sequencing performed for 45 matched cases of early-stage NSCLC.
Our findings align with those of prior research. Ni et al. conducted exome sequencing on single-cell CTCs from patients with lung cancer (14). Their findings revealed that single CTC harbored a greater number of mutations than primary cancer tissue, and the mutations in CTCs more closely resembled those in metastatic tumors than those in primary cancer tissue. Yoo et al. performed targeted deep sequencing on amplified DNA from CTCs in patients with lung cancer (16), reporting an absence of shared mutations between CTCs and primary cancer tissue. Furthermore, Kim et al. undertook whole exome sequencing in 20 patients with primary bladder cancers (17). Their research indicated no correlation in mutation abundance between primary cancer tissue and CTCs, and the concordance level of mutations was low.
Although the present results also indicated a low concordance rate between samples, our study methods require examination to confirm their validity, as the findings may impact future research on resectable NSCLC. First, we desired to ensure that the CTCs originated from the primary cancer. In our study, CTCs were detected in 24% of patients using immunofluorescence imaging, while 69% of patients exhibited mutated genes in CTCs. The mutational positivity of CTCs was relatively high for early-stage NSCLC. This high positivity rate could have resulted from the amplification and sequencing processes. However, our amplification method, REPLI-g, has demonstrated less bias and a higher yield compared to other methods (18). Furthermore, in another study, the positivity rate of CTCs in resectable early-stage NSCLC was reported to be around 60% using imaging methods (10), similar to our research. We believe that amplifying the DNA of CTCs allows for more sensitive detection than immunofluorescence imaging, especially considering the difficulty of detecting small quantities of CTCs in 10 cc of peripheral blood.
Second, we aimed to address some of the limitations associated with mutational studies of CTCs. Wang et al. suggested potential reasons for low concordance rates, such as the selection of subcolonies by the CTC isolation method and the reliance on EpCAM-dependent selection techniques (19). To encompass intra-tumor and CTC heterogeneity, we included gene mutations from various CTC colonies rather than from a single CTC. The EpCAM-dependent method tends to isolate only a limited number of CTCs (20), and EpCAM-positive CTCs have been found to be less frequently detected in NSCLC than in other epithelial tumors (21). Consequently, we employed a strategy that utilized multiple cell surface proteins, including EpCAM, EGFR, and HER2, which have been suggested to improve CTC isolation in NSCLC (21). We believe that this approach enables more specific detection of various CTCs, even in early-stage NSCLC, than other techniques. As an alternative, establishing CTC cultures could facilitate the expansion of CTCs to enough numbers for genotyping and phenotyping (17). However, this method may also be flawed if the cultured CTCs do not retain their original characteristics. Based on these considerations, we believe that our approach is reliable for the analysis of CTC mutations.
Multiple genes identified in CTCs were categorized to tumor suppressor genes, including MSH6, FANCE, CDK12, HNF1A, and AXIN2. Most of the genes are not directly associated with established mutational profiles of NSCLC (22). In contrast, all primary cancer tissue samples exhibited multiple mutations across both oncogenes (EGFR, NOTCH, ROS1, and KRAS) and tumor suppressor genes (TP53, FAT1, LRP1B, and PKHD1), which are well-known for their association with NSCLC. We applied Liang’s concept of “cancer hallmarks” to identify mutations specifically linked to carcinogenesis based on gene function (15). The CTCs showed a higher prevalence of genes involved in categories such as evasion of anti-growth signaling, replicative immortality, and invasion/metastasis. This indicates that primary cancer tissue likely arises from errors in proliferation or the cell cycle, while CTCs are more prone toward metastasis through invasion and replication, even in cases of early-stage NSCLC.
The potential reasons for the heterogeneity in mutational status between the two samples may include the influence of the microenvironment. Factors such as EMT, mesenchymal-to-epithelial transition, blood flow, and immune cell activity could impact the mutation of CTCs in a way markedly different from that of the primary tumor. Contrary to expectations, we found no correlation between the numbers or mutations of CTCs and pathologic stage or invasive pathologic factors. In our previous research, the immunophenotypes of captured CTCs exhibited a wide range, even among those at the same stage (23). Similarly, several studies have found that the number of CTCs does not vary significantly by stage in patients with lung cancer (12,24), a phenomenon observed in other types of cancer as well (25). We hypothesize that mutations in CTCs may occur independently of cancer stage or histologic type. These mutations could arise not because of cancer progression, but rather from the progression of the CTCs themselves, as indicated in Table 2. This suggests that certain mutations, such as those of MSH6 and FANCE, might be necessary for or associated with the differentiation of cancer cells into CTCs in NSCLC. However, this does not necessarily mean the mutations are critical for actual metastasis, since the CTCs collected using our specific markers did not precisely correspond to EMT. To elucidate the specific relationship between CTC mutations and metastasis, further studies that compare CTCs with actual metastatic tissue are needed.
To investigate which samples were more similar to advanced-stage NSCLC, we compared the mutations found in our study to those reported in advanced-stage NSCLC in other studies. Previous studies have also found EGFR, TP53, and KRAS as common mutations in advanced-stage NSCLC, similar to the mutations in primary cancer tissue in our study (26,27). However, because most prior studies performed panel sequencing that targeted fewer than 50 genes, we thought that it would not be appropriate to directly compare the mutations of advanced-stage NSCLC with those of CTCs in our study. Other studies utilizing panel sequencing with more than 50 genes have reported CDKN2A, PTCH1, and FLCN—which were detected in CTCs in our cohort—as among the top 10 genes (28,29). We hypothesize that advanced-stage NSCLC might have the characteristics of both primary cancer tissue and CTCs in early-stage NSCLC, and expect that it will be possible to detect more gene overlap between CTCs and advanced-stage NSCLC if the same 405-gene panel sequencing is used.
From a clinical perspective, we suggest that conducting a mutational analysis of CTCs can be useful at the time of preoperative evaluation. Patients with NSCLC who test positive for mutations observed in CTCs should be monitored for recurrence more closely during follow-up. If targetable mutations, such as EGFR, are detected in CTC samples, we should consider adjuvant systemic therapy after surgical resection. We can then determine whether to maintain or discontinue adjuvant therapy by conducting follow-up mutational analyses of CTCs. Several studies have suggested that targeted panel sequencing for the initial assessment of advanced-stage NSCLC could be cost-effective (30,31); however, the cost-effectiveness of mutational analysis of CTCs using panel sequencing in patients with early-stage NSCLC is doubtful.
Limitations
This study has several limitations. First, the small number of patients enrolled may not adequately represent the characteristics of primary NSCLC. Second, the follow-up duration for enrolled patients was too brief to establish an association between CTC mutation or positivity and cancer recurrence or survival. We plan to continue monitoring this cohort to understand long-term outcomes. Third, although the mutation profiles differed between the two samples as indicated in previous studies, we worried about differences between the concrete mutation profile of CTCs in this study and prior results. We are confident that the mutation profile of CTCs originated from the actual CTCs in the bloodstream and that their profile differed from that of the primary cancer tissue. However, due to the relatively early stage of the cohort in this study compared to the advanced or metastatic cohorts in previous research, minor mutations were detected more sensitively than the dominant mutations.
Conclusions
The mutational profiles of CTC DNA differed significantly from those of primary cancer tissue in early-stage NSCLC. Mutations in CTCs occurred in specific genes with high frequency and were associated with cancer metastasis, irrespective of stage or histological type. These findings contribute to a deeper understanding of CTCs in patients with early-stage NSCLC, however, further research with more cases is required.
Acknowledgments
Funding: This work was supported by the New Faculty Startup Fund from Seoul National University.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-709/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-709/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-709/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-709/coif). Jinsu Lee, Soojeong Ji and H.S. are from BeyondDx Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (approval No. B-2211-791-304) and informed consent was obtained from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- West HJ, Kim JY. Rapid Advances in Resectable Non-Small Cell Lung Cancer: A Narrative Review. JAMA Oncol 2024;10:249-55. [Crossref] [PubMed]
- Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, et al. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res 2016;5:466-82. [Crossref] [PubMed]
- Ren F, Fei Q, Qiu K, et al. Liquid biopsy techniques and lung cancer: diagnosis, monitoring and evaluation. J Exp Clin Cancer Res 2024;43:96. [Crossref] [PubMed]
- Luna J, Sotoca A, Fernández P, et al. Recent advances in early stage lung cancer. J Clin Transl Res 2021;7:163-74. [PubMed]
- Bogatsa E, Lazaridis G, Stivanaki C, et al. Neoadjuvant and Adjuvant Immunotherapy in Resectable NSCLC. Cancers (Basel) 2024;16:1619. [Crossref] [PubMed]
- Lin D, Shen L, Luo M, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther 2021;6:404. [Crossref] [PubMed]
- Jin F, Zhu L, Shao J, et al. Circulating tumour cells in patients with lung cancer universally indicate poor prognosis. Eur Respir Rev 2022;31:220151. [Crossref] [PubMed]
- Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. [Crossref] [PubMed]
- Frick MA, Feigenberg SJ, Jean-Baptiste SR, et al. Circulating Tumor Cells Are Associated with Recurrent Disease in Patients with Early-Stage Non-Small Cell Lung Cancer Treated with Stereotactic Body Radiotherapy. Clin Cancer Res 2020;26:2372-80. [Crossref] [PubMed]
- Fina E, Federico D, Novellis P, et al. Subpopulations of Circulating Cells with Morphological Features of Malignancy Are Preoperatively Detected and Have Differential Prognostic Significance in Non-Small Cell Lung Cancer. Cancers (Basel) 2021;13:4488. [Crossref] [PubMed]
- Moon SM, Kim JH, Kim SK, et al. Clinical Utility of Combined Circulating Tumor Cell and Circulating Tumor DNA Assays for Diagnosis of Primary Lung Cancer. Anticancer Res 2020;40:3435-44. [Crossref] [PubMed]
- Markou AN, Londra D, Stergiopoulou D, et al. Preoperative Mutational Analysis of Circulating Tumor Cells (CTCs) and Plasma-cfDNA Provides Complementary Information for Early Prediction of Relapse: A Pilot Study in Early-Stage Non-Small Cell Lung Cancer. Cancers (Basel) 2023;15:1877. [Crossref] [PubMed]
- Lyu M, Zhou J, Ning K, et al. The diagnostic value of circulating tumor cells and ctDNA for gene mutations in lung cancer. Onco Targets Ther 2019;12:2539-52. [Crossref] [PubMed]
- Ni X, Zhuo M, Su Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 2013;110:21083-8. [Crossref] [PubMed]
- Liang PI, Wang CC, Cheng HJ, et al. Curation of cancer hallmark-based genes and pathways for in silico characterization of chemical carcinogenesis. Database (Oxford) 2020;2020:baaa045. [Crossref] [PubMed]
- Yoo CE, Park JM, Moon HS, et al. Vertical Magnetic Separation of Circulating Tumor Cells for Somatic Genomic-Alteration Analysis in Lung Cancer Patients. Sci Rep 2016;6:37392. [Crossref] [PubMed]
- Kim TM, Yoo JS, Moon HW, et al. Distinct mutation profiles between primary bladder cancer and circulating tumor cells warrant the use of circulating tumors cells as cellular resource for mutation follow-up. BMC Cancer 2020;20:1203. [Crossref] [PubMed]
- Pinard R, de Winter A, Sarkis GJ, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 2006;7:216. [Crossref] [PubMed]
- Wang Q, Zhao L, Han L, et al. The Discordance of Gene Mutations between Circulating Tumor Cells and Primary/Metastatic Tumor. Mol Ther Oncolytics 2019;15:21-9. [Crossref] [PubMed]
- Satelli A, Batth I, Brownlee Z, et al. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget 2017;8:49329-37. [Crossref] [PubMed]
- Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Lee JM, Jung W, Yum S, et al. Prognostic Role of Circulating Tumor Cells in the Pulmonary Vein, Peripheral Blood, and Bone Marrow in Resectable Non-Small Cell Lung Cancer. J Chest Surg 2022;55:214-24. [Crossref] [PubMed]
- Wendel M, Bazhenova L, Boshuizen R, et al. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys Biol 2012;9:016005. [Crossref] [PubMed]
- Kong SL, Liu X, Suhaimi NM, et al. Molecular characterization of circulating colorectal tumor cells defines genetic signatures for individualized cancer care. Oncotarget 2017;8:68026-37. [Crossref] [PubMed]
- Fernandes MGO, Jacob M, Martins N, et al. Targeted Gene Next-Generation Sequencing Panel in Patients with Advanced Lung Adenocarcinoma: Paving the Way for Clinical Implementation. Cancers (Basel) 2019;11:1229. [Crossref] [PubMed]
- Pasquale R, Forgione L, Roma C, et al. Targeted sequencing analysis of cell-free DNA from metastatic non-small-cell lung cancer patients: clinical and biological implications. Transl Lung Cancer Res 2020;9:61-70. [Crossref] [PubMed]
- Zhou YJ, Zheng W, Zeng QH, et al. Targeted exome sequencing identifies mutational landscape in a cohort of 1500 Chinese patients with non-small cell lung carcinoma (NSCLC). Hum Genomics 2021;15:21. [Crossref] [PubMed]
- Lai WA, Huang YS, Chang KC, et al. Next-Generation Sequencing in Lung Cancers-A Single-Center Experience in Taiwan. Medicina (Kaunas) 2024;60:236. [Crossref] [PubMed]
- Steuten L, Goulart B, Meropol NJ, et al. Cost Effectiveness of Multigene Panel Sequencing for Patients With Advanced Non-Small-Cell Lung Cancer. JCO Clin Cancer Inform 2019;3:1-10. [Crossref] [PubMed]
- Abbass IM, Sheinson DM, Shah A, et al. Cost-effectiveness of large-panel next-generation sequencing in guiding first-line treatment decisions for patients with nonsquamous advanced non-small cell lung cancer. J Manag Care Spec Pharm 2024;30:649-59. [Crossref] [PubMed]


