
EGFR-V834L combined with L858R mutation reduced afatinib sensitivity and associated to early recurrence in lung cancer
Highlight box
Key findings
• We analyzed plasma cell-free DNA from 40 patients treated with afatinib as the first-line therapy. Epidermal growth factor receptor (EGFR)-V834L cooperates with other coexisting mutations to influence the therapeutic efficacy of EGFR tyrosine kinase inhibitors (EGFR-TKIs).
What is known and what is new?
• Non-small cell lung cancer with EGFR gene mutations has shown a dramatic response to EGFR-TKIs. However, recurrence due to acquired resistance is a problem in most cases.
• EGFR-V834L combined with L858R mutation reduced afatinib sensitivity and associated with poor progression-free survival.
What is the implication, and what should change now?
• It will be necessary to determine the types of co-mutations and clarify the role of these mutations in the efficacy of molecular-targeted drugs to develop better molecular-targeted therapies.
Introduction
Non-small cell lung cancer (NSCLC) with epidermal growth factor receptor (EGFR) gene mutations, such as exon 19 deletion (Ex19del) and L858R, has shown a dramatic response to EGFR tyrosine kinase inhibitors (EGFR-TKIs), leading to improved prognosis (1). To date, the first-generation reversible inhibitors gefitinib and erlotinib (2,3), the second-generation irreversible inhibitors afatinib and dacomitinib (4,5), and the third-generation osimertinib, which spares wild-type EGFR and is effective against the EGFR-T790M mutation (6), have been approved in many countries. However, recurrence due to acquired resistance is a problem in most cases (7).
Among these EGFR inhibitors, third-generation osimertinib is the most commonly used first-line treatment for EGFR-mutated NSCLC. However, there are no established treatments for patients with osimertinib resistance (8). The combination of first-generation EGFR-TKIs and vascular endothelial growth factor (VEGF)/VEGF receptor (VEGFR) inhibitors is considered a standard treatment, as the EGFR-T790M resistance mutation for first- and second-generation EGFR-TKIs is detected in approximately 50% of patients at the time of acquired resistance, and osimertinib is expected to be effective as a second-line therapy for such patients (9,10). In the National Comprehensive Cancer Network (NCCN) guidelines, the second-generation afatinib is also recommended as a first-line treatment for EGFR mutation-positive NSCLC (11). Afatinib shows a longer progression-free survival (PFS) as a first-line therapy than first-generation EGFR-TKIs; EGFR-T790M is detected in approximately 50% of patients at the time of acquired resistance (12) and osimertinib is expected to be effective as a second-line therapy. Clinical trials are currently underway to evaluate first-line therapy of afatinib followed by osimertinib for EGFR-mutated NSCLC (UMIN000037452) (13). In recent years, attention has been paid to the compound mutations that coexist with EGFR and affect sensitivity to EGFR-TKIs (14), and the efficacy of afatinib against compound mutations has also been reported (15). Based on these results, the importance of afatinib as a first-line therapy may increase, and it is necessary to elucidate the factors involved in the efficacy and resistance to afatinib to select subsequent treatments.
Analyses using clinical specimens are essential to elucidate the mechanisms of resistance. Tumor tissues are the gold standard, but tumor biopsy has several drawbacks, including invasiveness of the procedure and the inability to capture a complete picture of the tumor tissue due to intra-tumoral heterogeneity (16). Recently, liquid biopsy, which uses cell-free DNA (cfDNA) from blood, has attracted attention as a minimally invasive and easily reproducible biopsy method. Furthermore, because DNA from metastatic tumor cells in multiple organs enters the bloodstream, genetic alteration profiling using cfDNA may also reflect the heterogeneity of resistant tumor cells (17). Therefore, liquid biopsies are widely used in clinical practice.
The purpose of this study was to elucidate the gene alterations involved in the efficacy and resistance of afatinib by analyzing cfDNA obtained from patients with EGFR-mutated NSCLC treated with afatinib. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-471/rc).
Methods
Patients
This study was conducted as a prospective clinical trial in patients with EGFR-mutated NSCLC at Kanazawa University Hospital, Kanazawa Medical Center, Ishikawa Prefectural Hospital, Kouseiren Takaoka Hospital, Fukui-ken Saiseikai Hospital, Komatsu Municipal Hospital, Keiju Medical Center, Japanese Red Cross Kyoto Daini Hospital, Kyoto Prefectural University of Medicine, UJI-Tokushukai Medical Center, and Koseikai Takeda Hospital. The inclusion criteria were as follows: (I) patients aged 20 years or older at the time of registration; (II) patients who were histologically or cytologically diagnosed with NSCLC; (III) patients in whom EGFR mutations were detected in the tumor tissues at diagnosis; (IV) patients scheduled for afatinib monotherapy as the next-line treatment; (V) patients who were expected to survive for at least 3 months; and (VI) patients who provided written informed consent.
We collected demographic and clinical data of patients, including sex, smoking history, histological type of NSCLC, EGFR mutation type, treatment history for NSCLC prior to afatinib, as the clinical data at the initiation of afatinib, age, Eastern Cooperative Oncology Group performance status (ECOG PS), metastases sites, as the data about afatinib treatment, clinical response, PFS, time to treatment, afatinib dose reduction, and overall survival. Clinical responses were determined by clinicians at each institution according to the Response Evaluation Criteria in Solid Tumors version 1.1. PFS was defined as the date of commencing afatinib treatment until the earliest date of disease progression or death from any cause. Overall survival was determined from the date afatinib treatment commenced to the date of death from any cause.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Human Genome/Gene Analysis Research Ethics Committee of the Kanazawa University [approval No. #2016-039(434); UMIN000024476] and informed consent was obtained from all individual participants. The other institutions were also informed and agreed with the study.
Sample collections and cfDNA extractions
Peripheral venous blood (10 mL) was longitudinally collected in Cell-Free DNA BCT® (Streck Inc., La Vista, NE, USA) prior to afatinib treatment and at the time of resistance. Blood samples were sent to the laboratory of the Department of Respiratory Medicine, Kanazawa University Hospital, at room temperature within 3 days of collection. Immediately after arrival, plasma was collected by centrifugation, and cfDNA was extracted from the plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany). cfDNA was stored at −20 ℃ until use.
Analyses of gene alterations profile using cfDNA
Double-stranded cfDNA was quantified using a Qubit 2.0 Fluorometer and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Wilmington, NC, USA) according to the manufacturer’s instructions. Gene alteration profiling via deep sequencing was performed using the QIAseq Targeted DNA Panel (Human Lung Cancer Panel) (QIAGEN). This sequencing panel detects single point mutations, small insertions and/or deletions, and the amplicons are barcoded. A minimum of 10 ng cfDNA from each sample was used for library construction. The maximum attainable cfDNA was used for samples with low yields. All procedures were performed using the QIAseq Targeted DNA Panel kit, according to the manufacturer’s instructions.
QIAseq panel analysis was performed using the QIAGEN’s GeneGlobe data analysis portal. Somatic variants were distinguished from population polymorphisms by using the GnomAD database (http://gnomad.broadinstitute.org/). The clinical significant evidence and the clinical implications of variants were identified based on the annotations in public archives: ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), COSMICS (Catalogues of Somatic Mutations in Cancer; http://cancer.sanger.ac.uk/cosmic), and the Single Nucleotide Polymorphism Database (dbSNP). We included variants with a mapping quality ≥60, regardless of whether they have the ID on COSMICS or dbSNP. If the mapping quality ranged from 20 to 60, we adopted a variant with an ID in the public archives.
Cell lines
Ba/F3 cells were gifted by Dr. R. Katayama (Division of Experimental Chemotherapy, Cancer Chemotherapy Center, Japanese Foundation for Cancer Research, Tokyo, Japan), and were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS), 2 mmol/L glutamine, 1% penicillin-streptomycin (P/S), and mouse interleukin-3 (IL-3) (20 U/mL; Sigma-Aldrich, St. Louis, MO, USA). The Ba/F3 system is also available to investigate the sensitivity of driver mutants to therapeutic agents.
Establishment of a retroviral vector with random barcodes
The pCX6 vector was established by inserting random 10-base pair DNA barcode sequences upstream of the start codon of the corresponding genes into the pCX4 vector. Subsequently, full-length wild-type cfDNAs of the human EGFR and control genes were inserted into the pCX6 vector. Three EGFR variants were selected in this study. All plasmids which were encoding EGFR variants were constructed using the GeneArt Gene Synthesis System (Thermo Fisher Scientific) and sequenced using next-generation sequencing.
Creation of retrovirus and infection into cell lines
The recombinant plasmids were transduced with packaging plasmids (Takara Bio, Shiga, Japan) into HEK293T cells to generate recombinant retroviral particles. Ba/F3 cells were seeded on RetroNectin-coated (Takara Bio) 96-well plates and infected with retroviruses in RPMI 1640 medium supplemented with 20 U/mL IL-3. Individually transduced cells were collected and cultured to assess drug sensitivity.
Drug sensitivity assay in vitro
Cell viability was measured using a Cell Counting Kit-8 (CCK-8; DOJINDO, Kumamoto, Japan). Ba/F3 cells expressing EGFR variants were cultured in RPMI 1640 medium without IL-3. The transformed Ba/F3 cells that showed IL-3 independency were plated at a density of 2×103/100 µL per well in 96-well plates and were incubated for 24 hours, and then treated with the indicated concentrations of afatinib, osimertinib, and 10% dimethyl sulfoxide (as the vehicle control) for 72 hours. Cell growth was measured using the CCK-8 assay as described previously. The EGFR-TKIs used in the assays were purchased from Selleck Biotech (Yokohama, Japan). The experiment was conducted in both biological and technical triplicate.
Antibodies and western blot analysis
Antibodies against phosphorylated-EGFR (p-EGFR; Tyr1068; catalog 3777), total-EGFR (catalog 54359), p-extracellular signal-related kinase (p-ERK) (catalog 4370), total-ERK (catalog 4695), β-actin (catalog 3700), and cleaved caspase-3 (catalog 9661) were purchased from Cell Signaling Technology (Danvers, MA, USA) and diluted according to the manufacturer’s recommendations. Cells (2×105) were seeded in 6-well plates and incubated overnight before drug treatment for 48 h. Whole-cell lysates were prepared, and western blot was performed as described previously (18).
Tumor xenograft assay
We used 6-week-old male SHO-PrkdcscidHrhr mice (approval No. AP-173867, SHO-SCID mice from Charles River, Yokohama, Japan). Ba/F3 cells expressing EGFR variants were inoculated by subcutaneous injection of 3×106 cells in a 50/50 mixture of Matrigel and phosphate-buffered saline (PBS). One week after inoculation, mice were treated orally with the EGFR-TKIs, 2.5 mg/kg of afatinib, osimertinib (5 mg/kg), or vehicle control. The tumor volume was calculated using the following formula: π/6 × (large diameter) × (small diameter)2. Experiments were performed under a project license [No. #2016-039(434); UMIN000024476] granted by the Committee of Ethics of Experimental Animals and the Advanced Science Research Center, Kanazawa University (Kanazawa, Japan), in compliance with the Ministry of Education guidelines for the care and use of animals.
Statistical analysis
Survival analysis was performed using the Kaplan-Meier method. The Cox proportional hazards model was used for multivariable survival analysis, and variables with a P<0.1 in the univariate analysis were included in the multivariate analysis. Schoenfeld residuals were used to verify the proportional hazards assumption. All P values were two-sided, and P<0.05, unless otherwise stated, was considered statistically significant. All procedures were performed using R Studio. For data from the drug sensitivity assays in vitro and in vivo, the statistical significance of the differences was analyzed using a two-way unpaired t-test. All statistical analyses were performed using the GraphPad Prism ver. 9.0 (GraphPad Software, Inc., San Diego, CA, USA), with a two-sided P value.
Results
Patients and samples collected
A total of 55 patients were enrolled in the study. Afatinib treatment was discontinued in 49 patients and plasma samples were collected at the time of resistance in 40 patients. The reasons for discontinuation were disease progression in 38 patients and adverse events in two patients. One patient discontinued due to adverse event after beyond progressive disease (PD), and the other patient had disease progression after discontinuation due to adverse event. Thirty-five patients had experienced their dose reduction of Afatinib therapy and five patients had not. In 22 cases, paired samples were collected before afatinib treatment and at the time of acquisition of afatinib resistance. In 18 patients, samples were collected only during the acquisition of afatinib resistance (Figure 1).
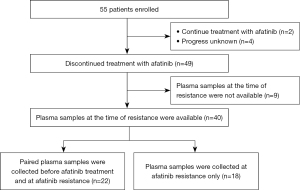
Analyses of cfDNA obtained before afatinib treatment from patients in which paired samples were collected
We analyzed plasma cfDNA from 22 patients whose paired plasma samples were collected before and after the acquisition of afatinib resistance. Of the 22 patients, 20 had adenocarcinoma and two had adenosquamous carcinoma (Table 1). EGFR-Ex19del, L858R, and G719X were detected in 10, eight, and four cases, respectively, by EGFR mutation analysis of the tumor tissues. In pretreatment plasma cfDNA, EGFR activating mutations were detected in 13 patients (59.1%) (Ex19del: seven cases, L858R: five cases, G719A: one case) (Figure 2). The most frequent coexisting mutations other than EGFR were KMT2D, LRP1B, and NF1, which were detected in 8 patients (38.1%). Among the 13 patients in whom EGFR driver mutations were detected in cfDNA, mutations that frequently coexisted included TP53, KMT2D, LRP1B, NF1, and SETD2 (five cases each, 38.5%). Interestingly, the EGFR-exon 21 V834L mutation was detected in two of the 13 cases in which EGFR-activating mutations were detected in the cfDNA (Figure 2). In both cases (Cases 1 and 2), V834L was present in the same allele (cis) as EGFR-L858R (Figure S1).
Table 1
| Characteristics | Data |
|---|---|
| Age (years) | 67.5 [39–77] |
| Gender | |
| Male | 8 (36.4) |
| Female | 14 (63.6) |
| Tumor histology | |
| Adenocarcinoma | 20 (90.9) |
| Adenosquamous carcinoma | 2 (9.1) |
| EGFR type | |
| Ex19del | 10 (45.5) |
| L858R | 8 (36.4) |
| Other† | 4 (18.2) |
| ECOG PS | |
| 0 | 5 (22.7) |
| 1 | 15 (68.2) |
| 2 | 2 (9.1) |
| Smoking | |
| Current or former | 10 (45.5) |
| Never | 12 (54.5) |
Data are presented as median [range] or n (%). †, G719X n=2, G719X+L861Q n=1, G719X+S768I n=1. EGFR, epidermal growth factor receptor; Ex19del, exon 19 deletion; ECOG PS, Eastern Cooperative Oncology Group performance status.
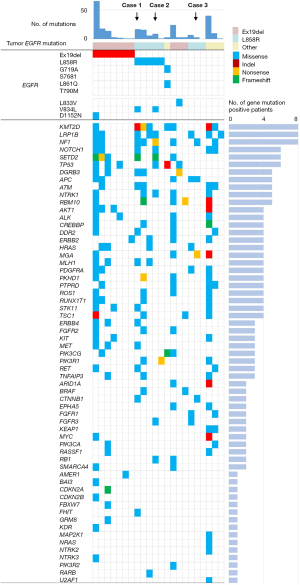
In the 22 patients, the median PFS (mPFS) after afatinib treatment was 9.1 months (Figure S2A). PFS was significantly shorter in patients with EGFR-L858R compared with that in patients with EGFR-Ex19del [mPFS: 10.7 vs. 4.5 months; hazard ratio (HR) 3.11; 95% confidence interval (CI): 0.99–9.70; P=0.051] (Figure S2B). Even when comparing patient backgrounds, no factors were found that could explain the poor prognosis in the L858R-positive group (Table S1). In these 22 patients, there was no significant correlation between the presence or absence of KMT2D, LRP1B, or NF1 mutations and PFS (Figure S3, Table S2). CDKN2A, CTNNB1, FGFR3, and RB1 mutations, which have been detected in cfDNA as genetic abnormalities associated with EGFR-TKI resistance (19-22), were found in the cfDNA of one, two, two, and two cases, respectively (Figure 2). Six patients with mutations in any of these genes in their cfDNA had significantly shorter PFS with afatinib than that of the 16 patients without these gene mutations (Figure S4).
In the 13 patients on whom EGFR driver mutations were detected, the PFS of the two cases with EGFR-L858R+V834L (Cases 1 and 2) was discernibly short (3.4 and 4.9 months). In the two cases of EGFR-L858R+V834L, coexisting mutations were found in TP53, CTNNB1, FGFR3, and RB1. We focused on this EGFR minor mutation; V834L and investigated the function.
Preclinical analyses to show the role of EGFR-V834L mutation on sensitivity to EGFR-TKIs
To test whether the EGFR-V834L mutation affects EGFR-TKI sensitivity, we performed functional genetic experiments by transfection of EGFR-L858R or L858R+V834L mutations in Ba/F3 cell lines. First, we assessed the sensitivity of the EGFR-TKIs afatinib and osimertinib. We observed increased cell viability of Ba/F3 cells transfected with EGFR-L858R+V834L in treatment with low concentrations of afatinib (0.3 nM) and osimertinib (10 nM), compared with that in those transfected with EGFR-L858R alone (Figure 3A). The difference was not statistically significant after treatment with EGFR-TKIs around the clinically available Cmax [Cmax: afatinib 26.3 nM (23), osimertinib 247 nM (24)] (Figure S5). Subsequently, we examined whether the addition of EGFR-V834L modulated signal transduction and cell apoptosis. As expected, the phosphorylation of EGFR and ERK was suppressed by treatment with a low concentration of afatinib in Ba/F3 cells transfected with EGFR-L858R but not in those transfected with EGFR-L858R+V834L (Figure 3B and Figure S6). In addition, the levels of cleaved caspase-3 were decreased in EGFR-L858R+V834L transfected cells compared with that of EGFR-L858R transfected cells. These results indicated that EGFR-V834L decreased the sensitivity of cells with EGFR-L858R to low concentrations of EGFR-TKIs in vitro.
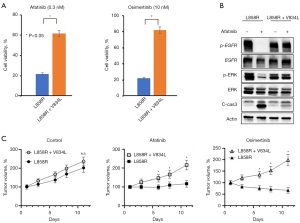
We further tested whether co-occurring EGFR-V834L in EGFR-L858R tumors contributed to EGFR inhibitor resistance in vivo. We treated mice bearing EGFR-L858R or EGFR-L858R+V834L tumor xenografts with afatinib, osimertinib, or vehicle and found that mice with EGFR-L858R+V834L tumors showed decreased afatinib and osimertinib sensitivity compared with that of mice bearing EGFR-L858R only tumors. We also confirmed that tumor growth was not impacted by EGFR-V834L addition in the vehicle treatment groups (Figure 3C). Collectively, these findings suggest that the addition of EGFR-V834L to EGFR-L858R affects sensitivity to EGFR-TKI treatment without a substantial impact on tumor cell growth.
Analyses of patients’ cfDNA obtained at afatinib resistance
Subsequently, we analyzed plasma cfDNA from 40 patients for whom specimens were collected during afatinib resistance (including 22 cases in which paired specimens were collected and 18 cases in which specimens were collected only during afatinib resistance). Of the 40 patients, 38 had adenocarcinoma and two had tumors of other histological types (Table S3). EGFR-Ex19del, L858R, and other mutations, including G719X, were detected in 22, 11, and seven cases, respectively, by EGFR mutation analysis of tumor tissues. In the plasma cfDNA obtained during afatinib resistance, EGFR driver mutations were detected in six cases (Ex19del: three cases, L858R: three cases) (Figure 4). EGFR-T790M mutation was detected in 2 of these patients (33%). EGFR-V834L was detected in two cases, both of which were EGFR-L858R positive. One of these cases was also positive for EGFR-V834L in cfDNA obtained before afatinib treatment (Case 2). The other case was negative for EGFR-L858R and V834L in cfDNA obtained before treatment, but positive for EGFR-L858R and V834L in cfDNA obtained during afatinib resistance (Case 3). EGFR-T790M was also detected in this case in cfDNA obtained after afatinib resistance. The cases in which EGFR-L858R and V834L were detected before afatinib treatment (Case 1) were negative for EGFR-L858R and V834 in the cfDNA obtained after afatinib resistance. This may be due to the low amount of tumor-derived DNA in the collected plasma samples.
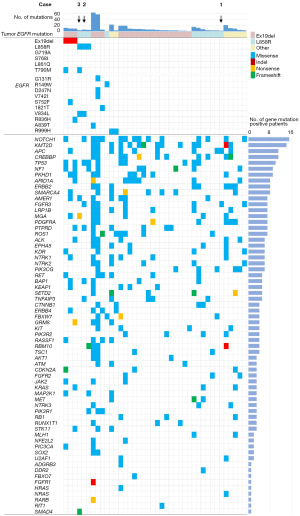
Among the 40 cases, the genes that were frequently co-mutated (>25%) were NOTCH1 (15 cases), KMT2D (14 cases), APC (11 cases), CREBBP (11 cases), and TP53 (10 cases) (Figure 4). Among the six cases in which driver EGFR mutations were detected in the cfDNA obtained during afatinib resistance, the most frequent co-mutations were TP53 (four cases) and NF1 (three cases). The mPFS of the 40 patients was 12.9 months. There was no significant difference on the PFS among the six cases in which driver EGFR mutations were detected in cfDNA at afatinib resistance and the 34 cases in which driver EGFR mutations were not detected in the cfDNA {8.9 months [95% CI: 4.13–not available (NA)] vs. 13.2 months (95% CI: 7.97–15.3); stratified HR, 1.50 (95% CI: 0.62–3.63); P=0.37} (Figure S7). In the first six cases, no correlation was observed between PFS and mutations in TP53 or NF1 (Figure S8).
In contrast, the mPFS of the three cases in which EGFR-V834L was detected in the cfDNA obtained before treatment or after afatinib resistance was clearly shorter (4.1 months) (Figure 5A). In the multivariate analysis, EGFR-V834L, CDKN2A, and CTNNB1 were independent factors for poor PFS (Table 2). When comparing PFS only in EGFR-L858R cases, although there was no statistically significant difference, the PFS of V834L positive cases tends to be shorter (Figure S9).
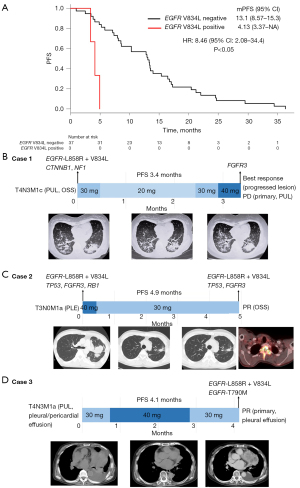
Table 2
| Characteristics | PFS (months) | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |||
| CDKN2A | ||||||||
| Negative | 13.1 | 1 | 1 | |||||
| Positive | 4.83 | 4.62 | 1.30–16.4 | 0.018 | 7.73 | 2.05–29.2 | 0.003 | |
| CTNNB1 | ||||||||
| Negative | 13.3 | 1 | 1 | |||||
| Positive | 4.13 | 4.44 | 1.71–11.6 | 0.002 | 4.13 | 1.39–12.2 | 0.01 | |
| EGFR-V834L | ||||||||
| Negative | 13.1 | 1 | 1 | |||||
| Positive | 4.13 | 8.46 | 2.08–34.4 | 0.003 | 6.04 | 1.27–28.8 | 0.02 | |
HR, hazard ratio; PFS, progression-free survival; CI, confidence interval; EGFR, epidermal growth factor receptor.
Case presentation of three patients with EGFR-L858R+V834L mutations
Finally, we present three cases in which EGFR-L858R+V834L was detected in the cfDNA.
Case 1
A 68-year-old woman, non-smoker, ECOG PS 1, had a primary tumor, intrapulmonary metastasis, and bone metastasis at the time of diagnosis; EGFR-L858R was detected in the primary tumor tissue by real-time PCR. Mutations in EGFR-L858R, EGFR-V834L, CTNNB1, and NF1 were detected in the cfDNA obtained before the first-line afatinib treatment. The PFS of afatinib treatment was 3.4 months, and the best response was PD. With afatinib resistance, the primary tumor and pulmonary metastases enlarged. The FGFR3 mutation, but not EGFR-L858R or EGFR-V834L, was detected in the cfDNA after afatinib resistance (Figure 5B).
Case 2
A 39-year-old man, who smoked, ECOG PS 1, had a primary tumor and pleural metastasis at the time of diagnosis. EGFR-L858R was detected in primary tumor tissue using real-time PCR. Mutations in EGFR-L858R, EGFR-V834L, TP53, FGFR3, and RB1 were detected in the cfDNA obtained before the first-line afatinib treatment. The PFS of afatinib treatment was 4.9 months, and the best response was partial response (PR). Bone metastases worsened with the development of afatinib resistance. Mutations in EGFR-L858R, EGFR-V834L, TP53, and FGFR3 were detected in the cfDNA after afatinib resistance (Figure 5C).
Case 3
A 74-year-old woman, non-smoker, ECOG PS 1, had a primary tumor in the left upper lobe, left malignant pleural effusion, malignant pericardial effusion, and intrapulmonary metastases at the time of diagnosis. EGFR-L858R was detected in primary tumor tissue using real-time PCR. Neither EGFR-L858R nor EGFR-V834L was detected in the cfDNA obtained before the first-line afatinib treatment. The PFS of afatinib treatment was 4.1 months, and the best response was PR. The primary tumor and cancerous pleural effusion worsened with the development of afatinib resistance. Mutations in EGFR-L858R, EGFR-V834L, and EGFR-T790M were detected in cfDNA during afatinib resistance (Figure 5D). It is possible that the driver EGFR-L858R and V834L mutations were not detected before treatment because the tumor burden (cfDNA amount) was small. It is also possible that EGFR-V834L induces afatinib tolerance in tumor cells and promotes acquisition of the EGFR-T790M mutation.
Discussion
We analyzed the plasma cfDNA of patients with EGFR-mutated lung cancer who received afatinib treatment as a first-line therapy and found that EGFR-V834L combined with EGFR-L858R was associated with short PFS after afatinib treatment. Furthermore, in vitro and in vivo analyses of gene transfer into Ba/F3 cells revealed that EGFR-V834L confers resistance of EGFR-L858R to low concentrations of EGFR-TKIs, including afatinib and osimertinib.
One of the issues with liquid biopsy is the low sensitivity for target gene alterations. In the present study, the positivity rate of driver EGFR mutations (Ex19del, L858R, and G719A) in plasma cfDNA before the first-line treatment was approximately 60%, a result similar to that of a previous report (25). In contrast, the number of positive results for driver EGFR mutations in the post-resistant plasma cfDNA was low (6/40 cases; approximately 20%). Nevertheless, EGFR-T790M was detected in two out of six cases (33%), and the detection rate by liquid biopsy was similar to that in a previous report (26). Therefore, the sensitivity for detecting gene mutations in this study was considered to be the same as previously reported. One of the reasons why the positivity rate of driver EGFR mutations in post-resistance samples was low may be the small tumor burden at the time of plasma sample collection. These findings highlight the importance of the timing of liquid biopsy specimen collection, which should be optimized in future studies.
In molecular structure, Ex19del removes from three to eight residues in the adenosine triphosphate (ATP)-binding site, while the L858R mutation is located away from the ATP-binding site, which is thought to result in different effects on EGFR-TKIs (27). The benefit of treatment with EGFR-TKI was more pronounced for patients with Ex19del compared to those with L858R mutations (28). In our study, PFS of patients with Ex19del was better than that of patients with L858R mutation. PFS of patients with L858R mutation was worse than previously reported (29). They may have had gene mutations that affect prognosis, and the small number of cases may have influenced the results of this study.
A study reported that V834 is located outside the ATP-binding pocket of EGFR (30), and because both valine and leucine are hydrophobic amino acids, it may not significantly affect protein structure. A recent report classified EGFR-L858R+V834L as a classical-like type mutation, which is located distal to the drug-binding pocket and has a modest to no impact on drug binding; however, detailed functional analysis has not been performed (31). On the other hand, it has been reported that V834L which results in a larger hydrophobic amino acid at V834 may result in steric hindrance of the drug near the anisole (methoxybenzene) group, and osimertinib may be less effective (32). Here, we introduced a genetic mutation into Ba/F3 cells and revealed, for the first time, that EGFR-V834L confers resistance to low concentrations of EGFR-TKIs in EGFR with the L858R mutation.
The LUX-Lung 2, 3, and 6 trials reported the efficacy of afatinib against uncommon mutations. However, there were no cases with co-mutations of L858R and V834L in the report. There is no comprehensive data on cases with co-mutations of L858R and V834L (15). Although a few papers reporting the incidence of V834L as a compound mutation in EGFR-mutated lung cancer, EGFR-L858R+V834L seems to be rare in NSCLC. To the best of our knowledge, only 12 cases of EGFR-L858R+V834L positive NSCLC have been reported, and V834L was selectively compounded with EGFR-L858R, and not EGFR-Ex19del, in all 12 cases (33-38). One report documented that EGFR-V834L could be detected as a germline mutation associated with familial lung cancer in tumors with EGFR-L858R+V834L mutations (33). It is not clear why V834L selectively coexists with EGFR-L858R rather than with EGFR-Ex19del.
In previous reports, the therapeutic efficacy of EGFR-TKIs in patients with NSCLC and EGFR-L858R+V834L varied. In one case, the PFS with erlotinib treatment was only 3 months for a 57-year-old man who was positive for the EGFR-V834L germline mutation. This finding indicated that erlotinib was not clinically effective in this case. In contrast, a 57-year-old woman, the older sister of the man in the previous case, with EGFR-V834L germline mutation and EGFR-L858R+V834L driver tumor mutation was treated with erlotinib for over a year, and EGFR-T790M was detected during resistance. Subsequently, osimertinib was administered, and EGFR-C797S was detected in the cis form of EGFR-T790M after 1 year of treatment (33). These observations indicated that erlotinib and osimertinib were clinically beneficial in this case. Furthermore, in the four cases reported by Wu et al. (38), the efficacy of EGFR-TKI treatment was PR in two cases and stable disease (SD) in two cases of NSCLC with EGFR-L858R+V834L. The results of these clinical reports and our preclinical experiments indicate that V834L confers resistance to low-concentration EGFR-TKIs in EGFR-L858R but may also affect the therapeutic efficacy of EGFR-TKIs by interacting with additional co-mutations. Studies have not clarified the types of co-mutations that coexist in patients with NSCLC having EGFR-L858R+V834L. In our study, genetic abnormalities associated with poor prognosis, such as TP53, CTNNB1, FGFR3, and RB1 mutations, which have been reported as poor prognostic factors in previous studies (20-22,39), coexisted. These factors may have acted together to shorten the PFS. In the future, it will be necessary to clarify the types of co-mutations that coexist with EGFR-L858R+V834L and the significance of these co-mutations in EGFR-TKI sensitivity to develop better molecular-targeted therapies.
This study had several limitations. First, the sample size was small. Another limitation was the sensitivity of the genetic analysis method used. We detected driver gene mutations in approximately 60% of plasma cfDNA collected before afatinib treatment of patients with EGFR-mutant lung cancer, which is similar to a previous report. However, more sensitive methods may be needed to discover novel biomarkers for predicting the efficacy of molecular target drugs. Moreover, the detection rate of driver EGFR mutations in cfDNA at afatinib resistance was 15%, suggesting that tumor cell-derived cfDNA may not have been sufficiently present in the plasma of many patients. To elucidate resistance mechanisms using cfDNA, it may be necessary to collect plasma samples multiple times when the tumor burden is relatively small.
Conclusions
We identified a V834L mutation coexisting with EGFR-L858R in patients with EGFR-mutated NSCLC having short PFS after first-line afatinib treatment. We found that the V834L mutation confers resistance to low-concentration EGFR-TKIs in EGFR-L858R-positive tumor cells, and it is possible that V834L cooperates with coexisting co-mutations to influence the therapeutic efficacy of EGFR-TKIs. In the future, in addition to compound mutations in driver genes, it will be necessary to determine the types of co-mutations and clarify the role of these mutations in the efficacy of molecular-targeted drugs to develop better molecular-targeted therapies.
Acknowledgments
We thank Ms. Miki Kashiwano (Department of Respiratory Medicine, Kanazawa University) for her technical assistance with cfDNA purification and Liu Yifeng (Department of Respiratory Medicine, Kanazawa University) for his additional experiments. We would also like to thank the patients, their families, and all the investigators who participated in this study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-471/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-471/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-471/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-471/coif). Tadaaki Yamada serves as an unpaid editorial board member of Translational Lung Cancer Research from October 2023 to September 2025. S.Y. obtained research grants from Chugai-Roche, AstraZeneca and Boehringer-Ingelheim, and speaking honoraria from Chugai-Roche, AstraZeneca and Boehringer-Ingelheim. Tadaaki Yamada obtained research grants from Ono Pharmaceutical, Janssen Pharmaceutical, AstraZeneca and Takeda Pharmaceutical Company Limited, and speaking honoraria from Eli Lilly, and Chugai-Roche. K.T. obtained research grants from Taiho Pharmaceutical, and speaking honoraria from Astrazeneca, Chugai-Roche, Eli-Lilly, MSD, Daiichi-Sankyo, Boehringer-Ingelheim and Ono Pharamaceutical. K.S. obtained speaking honoraria from AstraZeneca, Bristol Myers Squibb, and Chugai-Roche. Y.T. obtained research grants from AstraZeneca, Bristol Myers Squibb, Daiichi Sankyo, MSD, Beigene, and Regeneron. K.K. obtained research grants from Boehringer-Ingelheim. S.K. obtained research grants from Boehringer-Ingelheim. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Human Genome/Gene Analysis Research Ethics Committee of the Kanazawa University [approval No. #2016-039(434); UMIN000024476] and informed consent was obtained from all individual participants. The other institutions were also informed and agreed with the study. Experiments were performed under a project license [No. #2016-039(434); UMIN000024476] granted by the Committee of Ethics of Experimental Animals and the Advanced Science Research Center, Kanazawa University (Kanazawa, Japan), in compliance with the Ministry of Education guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer 2017;116:175-85. [Crossref] [PubMed]
- Ramalingam SS, O'Byrne K, Boyer M, et al. Dacomitinib versus erlotinib in patients with EGFR-mutated advanced nonsmall-cell lung cancer (NSCLC): pooled subset analyses from two randomized trials. Ann Oncol 2016;27:423-9. [Crossref] [PubMed]
- Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046-61. [Crossref] [PubMed]
- Suda K, Mizuuchi H, Maehara Y, et al. Acquired resistance mechanisms to tyrosine kinase inhibitors in lung cancer with activating epidermal growth factor receptor mutation--diversity, ductility, and destiny. Cancer Metastasis Rev 2012;31:807-14. [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]
- Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:1655-69. [Crossref] [PubMed]
- Laface C, Maselli FM, Santoro AN, et al. The Resistance to EGFR-TKIs in Non-Small Cell Lung Cancer: From Molecular Mechanisms to Clinical Application of New Therapeutic Strategies. Pharmaceutics 2023;15:1604. [Crossref] [PubMed]
- National Comprehensive Cancer Network. NCCN Guidelines Version 5.2024. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- Tanaka K, Nosaki K, Otsubo K, et al. Acquisition of the T790M resistance mutation during afatinib treatment in EGFR tyrosine kinase inhibitor-naïve patients with non-small cell lung cancer harboring EGFR mutations. Oncotarget 2017;8:68123-30. [Crossref] [PubMed]
- Morita R, Hata A, Ota T, et al. EP08. 02-133 Sequential Afatinib to Osimertinib in EGFR-mutant NSCLC: A Prospective Observational Study, Gio-Tag Japan Interim Report. J Thorac Oncol 2022;17:S466-7.
- Kohsaka S, Nagano M, Ueno T, et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med 2017;9:eaan6566. [Crossref] [PubMed]
- Yang JC, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Bedard PL, Hansen AR, Ratain MJ, et al. Tumour heterogeneity in the clinic. Nature 2013;501:355-64. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Taniguchi H, Akagi K, Dotsu Y, et al. Pan-HER inhibitors overcome lorlatinib resistance caused by NRG1/HER3 activation in ALK-rearranged lung cancer. Cancer Sci 2023;114:164-73. [Crossref] [PubMed]
- Jiang J, Gu Y, Liu J, et al. Coexistence of p16/CDKN2A homozygous deletions and activating EGFR mutations in lung adenocarcinoma patients signifies a poor response to EGFR-TKIs. Lung Cancer 2016;102:101-7. [Crossref] [PubMed]
- Zhou C, Li W, Shao J, et al. Analysis of the Clinicopathologic Characteristics of Lung Adenocarcinoma With CTNNB1 Mutation. Front Genet 2019;10:1367. [Crossref] [PubMed]
- Liao RG, Jung J, Tchaicha J, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res 2013;73:5195-205. [Crossref] [PubMed]
- Bhateja P, Chiu M, Wildey G, et al. Retinoblastoma mutation predicts poor outcomes in advanced non small cell lung cancer. Cancer Med 2019;8:1459-66. [Crossref] [PubMed]
- Murakami H, Tamura T, Takahashi T, et al. Phase I study of continuous afatinib (BIBW 2992) in patients with advanced non-small cell lung cancer after prior chemotherapy/erlotinib/gefitinib (LUX-Lung 4). Cancer Chemother Pharmacol 2012;69:891-9. [Crossref] [PubMed]
- Japan Package Insert. Accessed February 26, 2024. Available online: https://www.info.pmda.go.jp/go/pack/4291045F1027_1_12/
- Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta 2010;1804:559-66. [Crossref] [PubMed]
- Sheng M, Wang F, Zhao Y, et al. Comparison of clinical outcomes of patients with non-small-cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations after tyrosine kinase inhibitors treatment: a meta-analysis. Eur J Clin Pharmacol 2016;72:1-11. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Li M, Zhou CZ, Yang JJ, et al. The in cis compound EGFR mutations in Chinese advanced non-small cell lung cancer patients. Cancer Biol Ther 2019;20:1097-104. [Crossref] [PubMed]
- Robichaux JP, Le X, Vijayan RSK, et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021;597:732-7. [Crossref] [PubMed]
- Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195-203. [Crossref] [PubMed]
- van der Leest C, Wagner A, Pedrosa RM, et al. Novel EGFR V834L Germline Mutation Associated With Familial Lung Adenocarcinoma. JCO Precis Oncol 2018;2:PO.17.00266.
- Zhai SS, Yu H, Gu TT, et al. Lung adenocarcinoma harboring rare epidermal growth factor receptor L858R and V834L mutations treated with icotinib: A case report. World J Clin Cases 2020;8:3841-6. [Crossref] [PubMed]
- Li J, Zhang L, Wu Y, et al. Clinical Outcome and Molecular Analysis of a Chinese Patient With Lung Adenocarcinoma Harboring Rare EGFR Mutation V834L. J Thorac Oncol 2018;13:e189-91. [Crossref] [PubMed]
- Martin J, Lehmann A, Klauschen F, et al. Clinical Impact of Rare and Compound Mutations of Epidermal Growth Factor Receptor in Patients With Non-Small-Cell Lung Cancer. Clin Lung Cancer 2019;20:350-362.e4. [Crossref] [PubMed]
- Nie K, Jiang H, Zhang C, et al. Mutational Profiling of Non-Small-Cell Lung Cancer Resistant to Osimertinib Using Next-Generation Sequencing in Chinese Patients. Biomed Res Int 2018;2018:9010353. [Crossref] [PubMed]
- Wu SG, Chang YL, Yu CJ, et al. Lung adenocarcinoma patients of young age have lower EGFR mutation rate and poorer efficacy of EGFR tyrosine kinase inhibitors. ERJ Open Res 2017;3:00092-2016. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. J Clin Med 2020;9:1047. [Crossref] [PubMed]


