Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients
Introduction
The advent of genomic based precision medicine led to the implementation of biomarker testing in non-small cell lung cancer (NSCLC) patients. (1) In particular, EGFR mutation and ALK rearrangement assessments select patients for tyrosine kinase inhibitors (TKIs) administration (1,2). To date, tissue-based biomarker analysis still represents the gold standard; however, up to 30% of advanced NSCLC patients do not have tissue availability (3). In addition, testing for T790M mutation, at the time of progression to treatment with first or second generation TKIs is not always feasible on tissue (4-6). Due to tumor location or size, tissue biopsy can be risky. In these settings, liquid biopsy can represent a key opportunity. In particular, the EGFR mutational status can be assessed on circulating tumor DNA (ctDNA), even if it only represents a small fraction (<0.5%) of the cell free DNA (cfDNA) released into the blood (5,6). Most clinical trials adopted real time PCR (RT-PCR) or digital droplet PCR (ddPCR) (4-6) to compare the performance of EGFR testing in cfDNA versus tumor tissue. These “targeted methods” detect known mutations by specific probes, but do not cover the whole spectrum of EGFR mutations, missing the identification of less common but clinically relevant mutations. In addition, their multiplexing power is restricted, limiting the simultaneous analysis of additional emerging biomarkers. These limitations may be overcome by next generation sequencing (NGS), which enables the sequencing of large genomic regions or several exons on ctDNA (7-9). However, the requirement in this setting of a high degree of sensitivity may easily lead to false positive results, which underlines the need of a careful validation of a number of critical points, including blood collection, cfDNA extraction, library preparation, sequencing and variant calling (Figure 1). Here we review this evolving field, highlighting those methodological points that are crucial to accurately select NSCLC patients for TKIs treatment administration by NGS biomarker testing on cfDNA.

Liquid biopsy: cfDNA
In the targeted therapy glossary, liquid biopsies refer to those tests performed on body fluids aiming to predict patient treatment responses (4-6). In particular, whole blood has multiple sources that can be used for nucleic acid purification, including ctDNA (4-6). This represents a small fraction (<0.5%) of the total cfDNA passively released into the blood through necrosis or apoptosis of tumor cells or actively released by primary cancer cells or circulating tumor cells (CTCs) (4-9). Many clinical trials have intensively investigated the role of EGFR mutational assessment on ctDNA in NSCLC patients as an alternative to tumor tissue procurement; most of the data have been generated in patients in progressive disease to detect resistance mutations (e.g., T790M) (4-13).
NGS
NGS, based on massive parallel sequencing of millions of different DNA molecules, allows the detection of multiple mutations in multiple genes. By using focused gene panels the coverage can be narrowed down on clinically relevant targets so that each read is sequenced thousands of times, ensuring a high degree of sensitivity (8,11). As a general rule, this approach is named ultra-deep sequencing and enables to detect low abundant ctDNA in blood or other body fluids. Regardless from the platform used, NGS workflow is characterized by four steps: generation of a short fragment DNA library, single fragment clonal amplification, massive parallel sequencing and data analysis (7).
Critical workflow points: pre-analytical variables
Several preanalytical variables, such as blood collection and handling, cfDNA extraction protocols and storage temperature may affect the quantity and quality of cfDNA fragments in a sample. In this section, we analyze these points, suggesting the best option for clinical practice.
Blood collection and cfDNA extraction
In most clinical trials, EDTA containing tubes (Vacutainer) were used for blood collection (4-9). This approach avoids clotting and allows obtaining the plasma sample, that represent the matrix of choice for cfDNA extraction. Is particularly important, after the blood collection, to proceed with plasma preparation by centrifugation within 1 h, as ctDNA is associated with a high turn-over (15 min half-life). To overcome these problems, currently alternative blood collection tubes are under evaluation. In particular, PAXgene Blood DNA tubes (Qiagen) and Cell-Free DNA BCT tubes (STRECK) contain dedicated formaldehyde-free preservative agents that prevent the release of genomic DNA by inducing the stabilization of nucleated blood cells and the inhibition of the cfDNA degradation, enabling blood storage at room temperature. As underlined before, plasma remains the matrix of choice for cfDNA extraction, as the release of genomic DNA from white blood cells in the clotting process leads to a reduction of the ctDNA mutant allelic fraction in cfDNA in the case of serum. However, a few studies reported an improvement in sensitivity level between ctDNA and tissue based analysis for EGFR and BRAF mutational assessment by the concurrent analysis, in each given patient, of both serum and plasma derived cfDNA (6). In an ongoing study in a NGS Ampliseq based approach, we have determined that 9% of patients showed a concordant mutational status with tissues only in serum but not in plasma samples.
Considering these preliminary data, we suggest to analyze both serum and plasma derived cfDNA as a way to improve sensitivity in order to select patients eligible for TKI therapy.
Before cfDNA extraction, a pivotal point is also the time elapsing between blood collection and centrifugation. In addition, to keep the sample as informative as possible, pre-analytical handling of blood specimens is crucial. In our Institution special care is taken to standardize this process, by performing all the procedure in-house thanks to a dedicated nurse, who is a part of the laboratory staff. In our practice, for each patient a total of 10 mL of blood is collected in Vacutainer tubes (BD, Plymount, UK) to obtain 5mL of serum and 5 mL of plasma. After two centrifugation steps (2,300 rpm for 10 min) 1.2 mL of plasma and 1.2 mL of serum are used to extract cfDNA by using automatic platform (Qiagen, Venlo Limburg) following the manufacturer’s instructions. To remove any remaining cells, platelets, and cellular debris, other groups suggest the need for a third centrifugation step (1,000 g for 5 min) prior cfDNA extraction. Many kits, manual or automatic, have been developed to specifically extract cfDNA from plasma samples. The most commonly used in clinical trials was QIAamp circulating nucleic acid kit (Qiagen). Our group compared the performance of QIAamp circulating nucleic acid kit with that of the QIA symphony DSP Virus/Pathogen Midi Kit on the QIA symphony automatic platform. The results were similar, underling a possible advantage in term of consumable cost reduction, which may better allow for the implementation of testing not only on plasma, but also on serum. On the overall, to make cfDNA extraction in clinical practice more effective, we suggest the use of an automated procedure with a minimal input of 1 mL of serum and plasma, to obtain enough standardized cfDNA to use in subsequent library preparation process for NGS.
Critical workflow points: analytical variables
Library preparation and sequencing
The DNA to be sequenced is captured and used to construct a library, i.e., a collection of DNA fragments. When all of the known coding exons in a genome are captured, the approach is referred to as ‘whole exome sequencing’, which is usually used in research screens. In cytological applications, only a subset of the exome is amplified by a ‘targeted panel’. Several targeted capture panels are available, that usually cover 15–30 kb of genomic DNA to allow for the analysis of relevant key oncogenic mutational hotspots in 20–50 genes. Custom panels can be also designed by users.
Extracted cfDNA can be used to generate a collection of DNA fragments, that is generally termed as gene library (7,10). In particular, when all of the known coding exons in a genome are captured an ‘exome sequencing’ is obtained; this approach is usually carried out in research practice (7,10). Conversely, a target library composed by specific clinical relevant gene regions is obtained when narrow next generation panels are implemented (8,11). This latter approach better suits clinical practice and cfDNA applications, where a low limit of detection (LOD) is mandatory. Today, several targeted custom panels, covering relevant key oncogenic mutational hotspots in 20–50 genes (15–30 kb), are commercially available, but their limit of detection is not sufficient identifying somatic mutations in ctDNA (7). In our institution, we have developed and validated a custom target panel (5.2 kb) that covers clinically relevant hotspots in six different genes (EGFR, KRAS, NRAS, BRAF, cKIT and PDGFR) with a LOD of 0.02% of the mutated allele. This low LOD also reflects the library preparation approach used. Historically, the first approach, usually used by the Illumina (TM) platforms (San Diego, CA, USA), was based on hybridization capture system and involves the hybridization of DNA fragments, from a whole-genome preparation, to a mixture of probes designed with high specificity to match regions of a targeted panel of genes (7,10). This approach usually requires a starting DNA input of 250 ng, which may not be feasible in the ctDNA mutational assessment field; in addition, the hybridization–based capture system takes 24–72 h, which is too long for clinical purposes (10). More recently, a second library preparation modality, commonly used by Ion Torrent platforms (Life Technologies, Carlsbad, CA, USA), has been developed. It involves multiple primer pairs to make target capture directly by PCR (AmpliSeq kit, Lifetechnologies), requiring only a few (1-3) hours and a minimal DNA input (0.8 ng) (7).
Both library preparation approaches use fragments of synthetic DNAs with specific barcodes covalently added by DNA ligase to ensure simultaneous sequencing of different patient samples in the same run. Before sequencing reaction, to amplify the chemical signal, each single fragment of the library is clonally expanded in hundreds of thousands of copies (7,10). As a preliminary step, the library concentration is quantified, and any single DNA fragment of the library mixture is isolated, by the use of limiting dilution. In the Illumina platform, this reaction takes place on a solid support on a flat glass microfluidic channel (flow cell). Conversely, in the Ion Torrent Personal Genome Machine (PGM) the template is clonally amplified by an emulsion PCR on beads, which are singly trapped inside millions of oil micelles. In any case, the clonal amplification is digital in nature, as a single library fragment originates a single clone of amplified DNA, which in the following sequencing step will generate a single ‘read’, i.e., the sequence of nucleotides of each fragment being analyzed (10).
A key point for cfDNA analysis is the balance between the number of patients per run and the number of target regions amplified and sequenced for each patient. In fact, this can affect the amplicon coverage, reducing the reads depth. In our experience, by loading 16 sample patients (eight paired plasma and serum samples) onto a 6×106 wells chip (316 chip) with an 5.2 kb panel basing on AmpliSeq library preparation approach, a medium coverage of 2,000× for each amplicon can be achieved. This ultra-deep NGS approach, regardless the benchtop platform used, may represent the best option for the detection of low abundance ctDNA mutated alleles and enable the implementation of NGS in liquid biopsy clinical practice.
Variant calling and visual inspection
The combination of informatics tools used for processing, aligning and detecting variants in NGS data is usually termed as the bioinformatics pipelines. A plethora of informatics tools has been developed and are commercially available for tissue based analysis, but less is known regarding the appropriate parameter selection for ctDNA variant calling. First, any read is processed through filters that eliminate low-quality sequences. Then, the millions of reads produced need to be aligned to a reference human genome in order to generate interpretable sequencing results. Today, the human reference genome version currently used is 19th (hg19) (7-10); when differences occur between a base call and its aligned position to the hg19, variants are called. The implementation of different bioinformatics pipelines, either commercially available or developed in-house, can lead to a variability in the variant calling (7,8,11). Thus, the appropriate software settings should be optimized during the validation process taking into account the depth of coverage (number of reads covering a given base position), the average depth of coverage (average number of overlapping reads within the total sequenced area), the uniformity of coverage (distribution of coverage within specific targeted regions) and the specific parameter set for hotspots and non-hotspots variants (Table 1) (10). After the validation process, quality maintenance is challenging, since informatics is rapidly evolving, and software updates are frequent (10).

Full table
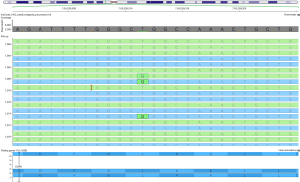
In some instances, the direct amplification for library preparation starting from low quantity cfDNA may increase the risk of strand bias (the ratio between the forward and reverse read), producing false negative results. To overcome this problem, following the clinicians’ request (e.g., evaluation of T790M mutation), visual inspection of produced reads using dedicated software, such as Integrated Genome Viewer or Golden Helix Genome Viewer, may help to define the mutational status of specific hotspot regions (Figure 2).
Conclusions
The evidences raised in this review showed that, after a careful validation process including blood collection, cfDNA extraction, library preparation, sequencing and variant calling, NGS can represent a new gold standard technique for cfDNA analysis in order to accurately select NSCLC patients for TKIs treatment administration. In addition, the simultaneously analysis of different variants in quantitative manner (e.g., EGFR exon 19 deletions and T790M point mutation) may help to define the time to switch from a first or second generation TKIs to a third generation TKIs therapy regimen.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn 2013;15:415-53. [Crossref] [PubMed]
- Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. [Crossref] [PubMed]
- Malapelle U, Pisapia P, Sgariglia R, et al. Less frequently mutated genes in colorectal cancer: evidences from next-generation sequencing of 653 routine cases. J Clin Pathol 2016;69:767-71. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Mead R, Duku M, Bhandari P, et al. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer 2011;105:239-45. [Crossref] [PubMed]
- Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 2012;30:1033-6. [Crossref] [PubMed]
- Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014;20:4613-24. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Chia PL, Do H, Morey A, et al. Temporal changes of EGFR mutations and T790M levels in tumour and plasma DNA following AZD9291 treatment. Lung Cancer 2016;98:29-32. [Crossref] [PubMed]