Blood-based biomarkers in lung cancer: prognosis and treatment decisions
Introduction
Outcomes and response to therapy of lung cancers can be highly variable. Tumor-tissue derived diagnosis has been the gold standard; and biomarkers derived from tumor tissue have been the main focus in establishing prognostic and predictive markers in lung cancer. However, reliance on tumor tissue has obvious shortcomings as it does not allow for serial sampling of tumor over the treatment course, nor does it allow for multiple biopsy attempts to assess intra-tumor heterogeneity. Therefore, the identification of novel biomarkers from blood to differentiate tumor from normal tissue, and to predict tumor behavior such as pathologic stage, response to chemotherapy or radiotherapy, and pattern of relapse, is of great importance in clinical practice. Blood-based biomarkers could capture the molecular diversity of the disease, while the ease of serial testing facilitates the monitoring of its spatial and temporal progression.
Circulating tumor cells (CTCs)
CTCs are shed from the primary tumor into the circulation and are considered to contribute to cancer progression (1). Tumor-derived cells identified in peripheral blood can be used for diagnosis of original cancer or detection of recurrent cancer (“liquid biopsy”) (2). This topic will be covered by a separate review and will not be elaborated upon here.
To date, multiple techniques have been developed for the isolation, characterization, and enumeration of CTCs. These methods present great variability in CTC detection rates, sensitivity, and specificity (3). Commonly, CTC assays utilize an initial enrichment step with epithelial markers to deplete blood-derived cell populations and a detection step to identify CTCs. However, the rarity of CTCs, the lack of a consensus definition on biomarker expression in CTCs, and the significant inter- and intra-patient heterogeneity of these cells, represent significant challenges in CTC detection. To date, the CellSearch (Veridex) system is currently the only US FDA approved technology for this purpose (4-6). CellSearch (Veridex) employs an epithelial cell adhesion molecule (EpCAM)-based enrichment step and identifies CTCs as intact enucleated cells staining positive for cytokeratins and negative for the leucocyte marker CD45 (7).
The presence of CTCs has been associated with poor outcomes in advanced stage cancer patients (8-10). With the use of the CellSearch system, CTC number was a strong predictor of overall survival (OS) in a cohort of 101 patients with non-small cell lung cancer (NSCLC) (11). Baseline CTC counts of over 5/7.5 mL were associated with shorter progression free survival (PFS) and OS in patients with NSCLC (11). The prognostic role of CTC detection has also been shown in surgically resected NSCLC (10). Furthermore, the prognostic significance of CTCs in lung cancer was demonstrated in a meta-analysis of 20 trials comprising 1,576 NSCLC patients, which revealed a significant association between the presence of CTCs and decreased disease-free survival (DFS) and decreased OS (12). Despite these results, the use of CTCs as a prognostic biomarker in NSCLC has not been routinely utilized clinically.
Circulating (cell-free) tumor DNA (ctDNA)
Lung cancer cells release DNA fragments into the circulation, and these fragments can be found in the cell-free fraction of blood together with DNA fragments from normal cells (ctDNA). It has been speculated that ctDNA is released passively into the bloodstream from dead/dying tumor cells, a process related to the tumor burden, tumor growth, and anti-tumor therapy. While the presence of ctDNA has long been recognized, techniques that are sensitive and specific enough to detect it have only become available recently. These techniques include BEAMing (beads, emulsion, amplification, magnetics) technology and CAPP-seq (cancer personalized profiling by deep sequencing) among others.
The ctDNA levels have been consistently shown to be higher in NSCLC patients compared with healthy control subjects (13,14). A large body of evidence has demonstrated the prognostic significance of ctDNA levels. It has been shown that higher pretreatment ctDNA levels were associated with worse OS in patients who were treated with first-line platinum doublet chemotherapy (16.8 vs. 22.4 months) (15). In another report, increased plasma ctDNA levels were correlated with advanced tumor stage, tumor progression after chemotherapy as well as poor survival (16,17).
A recent report utilized CAPP-Seq ctDNA analysis to study resistance mechanisms in 43 lung cancer patients treated with the third-generation EGFR inhibitor rociletinib (18). Multiple resistance mechanisms have been recognized in 46% of patients after treatment with first-line inhibitors, indicating frequent intra-patient heterogeneity. In a separate report, ctDNA was detected in 100% of patients with stage II–IV NSCLC and in 50% of patients with stage I disease, indicating its prognostic value and the specificity was 96% (19). Furthermore, levels of ctDNA were highly correlated with tumor volume and distinguished between residual disease and treatment-related imaging changes. This suggests that the measurement of ctDNA levels would allow for earlier response assessment than radiographic approaches.
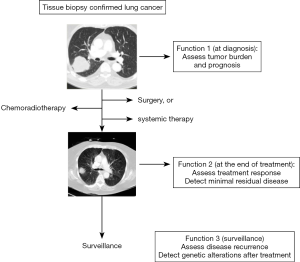
Due to the ease of serial sample collections from patients during cancer surveillance or treatment, and the development of robust technology to detect driver mutations from small amount of ctDNA, ctDNA analysis has become very useful in lung cancer monitoring and guiding treatment decisions. For example, a patient who had been treated with erlotinib for a lung cancer with EGFR exon 19 deletion, could later develop a mutation in the EGFR gene which confers resistance to Erlotinib. Obtaining blood samples at the time of tumor recurrence or progression followed by EGFR mutation analysis, would help the clinicians to decide if the tumor has acquired a mutation that led to resistance to a first-generation EGFR inhibitor. Other functions of ctDNA include monitoring minimal residual disease as well as possible tumor recurrence (Figure 1) (10).
MicroRNAs (miRs)
miRs are small noncoding single stranded RNAs with a length of 19 to 22 nucleotides. They function by silencing mRNA, thus negatively regulating gene expression at the post transcriptional level (20). The role of miR expression in tumor samples has been widely reported, but data on the prognostic value of circulating miR has been scarce.
A serum miR signature of miR-486, miR-30d, miR-1, and miR-499 has been shown to be an independent predictor of survival in NSCLC patients (21). In a separate report, high miR-125b serum expression was shown to be an independent predictor of poor survival in an analysis of 193 NSCLC patients of various stages (22). A signature of 5 miR gene set has recently been identified by Silva et al. as a potential prognostic marker, and was validated in a separate cohort of 78 NSCLC patients and 48 control participants. In the same study, the levels of let-7f and miR-30e-3p have been shown to correlate with poor outcomes (23).
Exosomes are membrane-enclosed extracellular vesicles (EVs) with a diameter of 30–100 nm (24) that are derived from endosomes (25). All cell types can secrete exosomes and the formation and release of exosomes are highly regulated. Cancer cells are known to secrete more exosomes. Exosomes are stable and can be isolated from diverse body fluids, including plasma, serum and urine. Moreover, RNA, including miRs are much more stable when isolated and stored together with exosomes, providing higher yield. In a recent study (26), with a quantitative polymerase chain reaction (qPCR) array panel, 84 plasma exosomal miR samples were analyzed from 10 patients with NSCLC and 10 healthy controls. Elevated levels of exosomal miR-23b-3p, miR-10b-5p and miR-21-5p were independently associated with poor OS [with hazard ratio of 2.42 (1.45–4.04), P=0.001; 2.22 (1.18–4.16), P=0.013; 2.12 (1.28–3.49), P=0.003, respectively]. When compared to a model derived from clinical prognostic variables, adding the three exosomal miRNA signatures significantly improved survival predictive accuracy with an increase of time-dependent area under the receiver operating characteristic curve from 0.88 to 0.91 (P=0.015) (26).
miRs are useful not only as diagnostic biomarkers but also as potential prognostic and predictive markers. For example, increased miR-21 leads to in vitro platinum resistance whereas the expression of miR-21 in tumor tissue and plasma was associated with DFS after adjuvant platinum-based chemotherapy (27). Upregulation of circulating miR-22 levels was found to predict resistance to pemetrexed in patients with advanced NSCLC (28).
Other blood-based biomarkers
Many attempts have been made to exploit serum or plasma biomarkers, including proteomic or genomic features, in order to identify prognostic and/or predictive classifiers for lung cancer patients. For example, VeriStrat®, a serum or plasma-based test which was developed using matrix-assisted laser desorption/ionization (MALDI) mass spectrometry, has been shown to be prognostic for both OS and PFS, independent of clinical features. The test is highly predictive of objective response to erlotinib (29) or combination TKI treatment (30). The VeriStrat algorithm has been interrogated retrospectively and prospectively in samples from several randomized trials, such as BR21, confirming the prognostic information associated with the molecular signature.
Numerous other serum/plasma based tests including DNA methylation signatures in circulating DNA (31), and serum autoantibodies against tumor-associated antigens (TAAs) (32), have shown some promise as potential early detection tools or prognostic indicators. However, as these tests generally lack specificity, more robust biomarkers have been identified and incorporated gradually into clinical practice (such as liquid biopsy tests).
Conclusions
Circulating tumor markers can be useful tools for predicting outcomes and treatment response in lung cancer patients. Some have already entered routine clinical practice. The integration of prospective biomarker analysis and imaging studies in randomized clinical trials will make it possible to validate the utility of blood-based tumor biomarkers in lung cancer diagnosis, prognosis and research. Successes in this field of endeavor will presumably encourage future efforts to identify and validate new biomarkers.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. [Crossref] [PubMed]
- Hanssen A, Loges S, Pantel K, et al. Detection of circulating tumor cells in non-small cell lung cancer. Front Oncol 2015;5:207. [Crossref] [PubMed]
- Xenidis N, Perraki M, Kafousi M, et al. Predictive and prognostic value of peripheral blood cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in node-negative breast cancer patients. J Clin Oncol 2006;24:3756-62. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [Crossref] [PubMed]
- Mocellin S, Hoon D, Ambrosi A, et al. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res 2006;12:4605-13. [Crossref] [PubMed]
- Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539-46. [PubMed]
- Pérez-Callejo D, Romero A, Provencio M, et al. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res 2016;5:455-65. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One 2013;8:e78070. [Crossref] [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. [Crossref] [PubMed]
- Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer 2009;64:92-7. [Crossref] [PubMed]
- Catarino R, Coelho A, Araújo A, et al. Circulating DNA: diagnostic tool and predictive marker for overall survival of NSCLC patients. PLoS One 2012;7:e38559. [Crossref] [PubMed]
- Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic Acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol 2004;22:4157-64. [Crossref] [PubMed]
- Nygaard AD, Holdgaard PC, Spindler KL, et al. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 2014;110:363-8. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. [Crossref] [PubMed]
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 2010;28:1721-6. [Crossref] [PubMed]
- Yuxia M, Zhennan T, Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol 2012;138:2045-50. [Crossref] [PubMed]
- Silva J, García V, Zaballos Á, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J 2011;37:617-23. [Crossref] [PubMed]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [PubMed]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83. [Crossref] [PubMed]
- Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017;8:13048-58. [PubMed]
- Gao W, Lu X, Liu L, et al. MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther 2012;13:330-40. [Crossref] [PubMed]
- Franchina T, Amodeo V, Bronte G, et al. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol 2014;229:97-9. [PubMed]
- Carbone DP, Ding K, Roder H, et al. Prognostic and predictive role of the VeriStrat plasma test in patients with advanced non-small-cell lung cancer treated with erlotinib or placebo in the NCIC Clinical Trials Group BR.21 trial. J Thorac Oncol 2012;7:1653-60. [Crossref] [PubMed]
- Kuiper JL, Lind JS, Groen HJ, et al. VeriStrat(®) has prognostic value in advanced stage NSCLC patients treated with erlotinib and sorafenib. Br J Cancer 2012;107:1820-5. [Crossref] [PubMed]
- Lissa D, Robles AI. Methylation analyses in liquid biopsy. Transl Lung Cancer Res 2016;5:492-504. [Crossref] [PubMed]
- Dai L, Tsay JC, Li J, et al. Autoantibodies against tumor-associated antigens in the early detection of lung cancer. Lung Cancer 2016;99:172-9. [Crossref] [PubMed]