Inhibition of MEK, a canonical KRAS pathway effector in KRAS mutant NSCLC
KRAS mutant NSCLC cells require active nuclear export of Iκβα (also known as NFKBIA), a negative regulatory protein of NF-κB signaling, for maintaining survival signaling (1-3). Nuclear export receptor XPO1 correlates with KRAS mutation status. Sensitivity to XPO1 inhibitors (KPT-330 or Selinexor) is associated with apoptosis in KRAS mutant cell lines. In contrast, chemical inhibition of mitogen-activated protein kinase kinase (also known as MEK) has little consequence on cell viability (1). XPO1 inhibitors induce the nuclear accumulation of Iκβα in a broad panel of tested cell lines, indicating that selective sensitivity is related to inhibition of NF-κB signaling (4). Jänne et al. (5) carried out the phase 3 Selumetinib Evaluation as Combination Therapy (SELECT-1) trial which assessed second line selumetinib plus docetaxel for patients with KRAS mutant, metastatic NSCLC versus placebo plus docetaxel. The SELECT-1 trial did not improve progression free survival (PFS) or overall survival (OS). Median PFS was 3.9 months in the selumetinib plus docetaxel group and 2.8 months in the placebo plus docetaxel group. Median OS was 8.7 months in the selumetinib plus docetaxel group versus 7.9 months in the placebo plus docetaxel group. The Jänne et al. study highlights many aspects of the difficulties in treating KRAS mutant NSCLC patients. The meager effect of selumetinib as a MEK inhibitor should be revisited based upon the abundant information reaped from the study to move forward from bench to bed. Undeniably, there are multiple approaches. Firstly, KRAS protein induced XPO1-dependent activation of NF-κB signaling in NSCLC cells (1) should be explored. This activation is not required for wild-type tumor NSCLC lines and XPO1 inhibitors warrant testing in the clinical setting. Noteworthy is the fact that FSTL5 mutations found in KRAS mutant cell lines were resistant to XPO1 inhibitors. Somatic mutations in FSTL5 are found in 10% of lung adenocarcinomas. FSTL5 depletion produces sensitivity to XPO1 inhibitors in KRAS mutant, FSTL5 wild-type NSCLC cell lines. Notably, FSTL5 depletion induces YAP1 activation, akin to that induced upon depletion of the LATS1 and LATS2 tumor suppressor genes (1). There is strong evidence between the FSTL5 mutation status and YAP1 protein accumulation. Intriguingly, we show that an increase in YAP1 in BRAF and KRAS mutant NSCLC tumors is a biomarker predicting worse response to RAF and MEK inhibition in patients (6). Secondly, it has been reported that the IκB kinase (IKK)-related kinases TANK-binding kinase-1 (TBK1) and IKKε promote KRAS driven activity by regulating interleukin (IL)-6 and identify CYT387 as a potent JAK/TBK1/IKKε inhibitor (7). Thirdly, MEK inhibitors are clinically active in BRAFv600E mutant melanomas, but only marginally active in KRAS mutant tumors. MEK inhibitors induce RAF-MEK complexes in KRAS mutant models and disrupting such complexes enhanced inhibition of RAF proto-oncogene serine/threonine-protein kinase (CRAF)—dependent extracellular signal-regulated kinase (ERK) signaling (8). In fact, ablation of CRAF expression induces regression of KRAS-Trp53 mutant lung tumors (9). The combination of sorafenib [a multi-kinase inhibitor that targets both, CRAF and BRAF, as well as vascular endothelial growth factor receptor (VEGFR)] and aspirin in KRAS mutant NSCLC cells produces a significant reduction of cell proliferation within 72 hours in A549 and H358 cells by simultaneously effecting two independent pathways when the tumor cells were sensitive to single agents, sorafenib and aspirin (10). Although trametinib is superior to other MEK inhibitors since it impairs feedback reactivation of ERK, it activates multiple signaling pathways, reflecting a relief in feedback mechanisms produced by hyperactive KRAS signaling in KRAS mutant NSCLC cells (11,12). Trametinib, as other MEK inhibitors, activates signal transducer and activator of transcription 3 (STAT3), as well as several receptor tyrosine kinases (RTKs), including fibroblast growth factor receptor 1 (FGFR1) and the FGFR adaptor protein, fibroblast growth factor receptor substrate 2 (FRS2) (11,13). The sensitivity to the combination of trametinib and FGFR inhibition (ponatinib) correlates with the degree of FRS2 phosphorylation after trametinib treatment (11). Intriguingly, in combination with trametinib, afatinib shows activity in KRAS mutant NSCLC lines (11) in accordance with other findings that epithelial KRAS mutant NSCLC cell lines overexpress ERBB3 and are sensitive to the combination of afatinib plus a MEK inhibitor, while mesenchymal KRAS mutant NSCLC cell lines following MEK inhibition overexpress FGFR1 and FRS2, and, henceforth, are sensitive to the combination of a MEK inhibitor plus an FGFR inhibitor (NVP-BGJ398) (14). The fact that activation of YAP1 stimulates secretion of FGF ligands and expression of FGFR in ovarian cancer is significant (15). Different lines of evidence show that, following MEK inhibition, there could be overexpression of other RTKs, like MET and AXL, as well as overactivation of Src-YAP1-NOTCH-HES1, in addition to STAT3 (16,17). AXL overexpression has been a trait of KRAS mutant cell lines with mesenchymal features responding to the combination of erlotinib and an AXL inhibitor (18), or the combination of the AXL inhibitor, TP0903, plus a PARP inhibitor (olaparib) (19). Inhibition of AXL directly reverts the epithelial-mesenchymal transition (EMT) phenotype and leads to decreased expression of DNA repair genes, diminishing homologous recombination proficiency (19). The combination of a WEE1 inhibitor with an MTOR inhibitor has been reported in mutant KRAS NSCLC tumors (20). The combination of MEK inhibitors with Src inhibitors could be of great interest, since a transmembrane protein, CUB domain-containing proteins (CDCP1), is required for the functional link between RAS and Src signaling. Most KRAS mutant NSCLC tumors overexpress CDCP1 (21). CDCP1 can also interact with and activate all Src-family kinase (SFK) members, such as, YES and LYN (17,22). At least 21% of c NSCLCs show significant integrin β3 (ITGB3) mRNA expression and targeting galectin-3 could be a novel strategy for such KRAS mutant tumors addicted to integrin αvβ3/galectin-3 (GCS-100) (23).
Loss of function of MutT homolog 1 (MTH1), a nucleotide pool sanitizing enzyme, impairs growth of KRAS mutant tumor cells. Overexpression of MTH1 mRNA levels has been shown to be a prognostic factor, documented in lung cancer and renal cell carcinoma, and MTH1 inhibitors are in development. It was found that (S)-crizotinib efficiently inhibited colony formation of KRAS mutated cells, like an MTH1 inhibitor (SCH51344). (S)-crizotinib is less potent than the (R)-enantiomer against the established anaplastic lymphoma kinase (ALK), MET and ROS1 (24).
Justilien and Fields describe the relevance of protein kinase Cι (PKCι) in KRAS mutant NSCLC, activating a RAC1-PAK-MEK1,2-ERK1,2 signaling pathway and show that epithelial cell transforming sequence 2 (Ect2), a guanine nucleotide exchange factor for Rho family GTPases is amplified and overexpressed with PKCι in NSCLC tumors (25). Justilien has also proven relevant that Ect2 is required for KRAS-Trp53 lung tumorigenesis (26), as well as the fact that PKCι activates NOTCH3 signaling (27). The studies of Justilien and Fields demonstrate that auranofin (a PKCι inhibitor) could be cardinal for treatment (28) and combinations of auranofin with PAK inhibitors deserve further testing (16) (Figure 1).
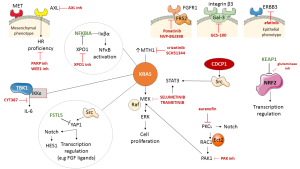
Finally, KEAP1 mutations are frequent in NSCLC, with KRAS mutant NSCLC accounting for 20%. The KEAP1 gene encodes Kelch-like ECH-associated protein 1, a negative regulator of nuclear factor erythroid 2-like 2 (NFE2L2; NRF2) (29). KRAS mutant cell lines carrying KEAP1 mutations are sensitive to glutaminase inhibition since such cell lines are dependent upon glutaminolysis. Furthermore, NRF2 is a master transcriptional regulator that confers chemo-resistance. The clinical outcomes of the SELECT-1 study highlight the limited effect of current therapeutic approaches either with chemotherapy or MEK inhibitors in KRAS mutant NSCLC. The Jänne et al. study openly shows the dismal outcome of NSCLC patients with KRAS mutations and therapeutic solutions should be urgently developed for more molecularly individualized clinical trial models, as is common in breast cancer, such as the My Pathway trial (30). Figure 1 illustrates several layers of research, including potential biomarkers involving pathways and intercommunication between different components, from RTKs on the cell surface, to the cytoplasm and nuclear components of the tumor cells. Importantly, selective inhibition of MET can lead to overexpression of FRS2 and the combination with FGFR inhibitors is warranted, particularly in mesenchymal tumors displaying elevated expression of AXL. Other opportunities are also depicted in Figure 1 and the accumulated evidence described herein can help pave the way for better therapies in KRAS mutant NSCLC patients.
Acknowledgements
Funding: Work in Dr. Rosell’s laboratory is partially supported by a grant from La Caixa Foundation, an Instituto de Salud Carlos III grant (RESPONSE, PIE16/00011) and a Marie Skłodowska-Curie Innovative Training Networks European Grant (ELBA No. 765492). Dr. Masaoki Ito’s work has been funded by the IASLC Fellowship and Young Investigator Award.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Kim J, McMillan E, Kim HS, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature 2016;538:114-7. [Crossref] [PubMed]
- Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer 2013;14:205-14. [Crossref] [PubMed]
- Rosell R, Gonzalez-Larriba JL, Alberola V, et al. Single-agent paclitaxel by 3-hour infusion in the treatment of non-small cell lung cancer: links between p53 and K-ras gene status and chemosensitivity. Semin Oncol 1995;22:12-8. [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Jänne PA, van den Heuvel MM, Barlesi F, et al. Selumetinib Plus Docetaxel Compared With Docetaxel Alone and Progression-Free Survival in Patients With KRAS-Mutant Advanced Non-Small Cell Lung Cancer: The SELECT-1 Randomized Clinical Trial. JAMA 2017;317:1844-53. [Crossref] [PubMed]
- Lin L, Sabnis AJ, Chan E, et al. The Hippo effector YAP promotes resistance to RAF-and MEK-targeted cancer therapies. Nat Genet 2015;47:250-6. [Crossref] [PubMed]
- Zhu Z, Aref AR, Cohoon TJ, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov 2014;4:452-65. [Crossref] [PubMed]
- Lito P, Saborowski A, Yue J, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 2014;25:697-710. [Crossref] [PubMed]
- Sanclemente M, Francoz S, Esteban-Burgos L, et al. c-RAF Ablation Induces Regression of Advanced Kras/Trp53 Mutant Lung Adenocarcinomas by a Mechanism Independent of MAPK Signaling. Cancer Cell 2018;33:217-28.e4. [Crossref] [PubMed]
- Hammerlindl H, Ravindran Menon D, Hammerlindl S, et al. Acetylsalicylic Acid Governs the Effect of Sorafenib in RAS-Mutant Cancers. Clin Cancer Res 2018;24:1090-102. [Crossref] [PubMed]
- Manchado E, Weissmueller S. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016;534:647-51. [Crossref] [PubMed]
- Rosell R, Karachaliou N, Morales-Espinosa D, et al. Adaptive resistance to targeted therapies in cancer. Transl Lung Cancer Res 2013;2:152-9. [PubMed]
- Lee HJ, Zhuang G, Cao Y, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 2014;26:207-21. [Crossref] [PubMed]
- Kitai H, Ebi H, Tomida S, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov 2016;6:754-69. [Crossref] [PubMed]
- Hua G, Lv X, He C, et al. YAP induces high-grade serous carcinoma in fallopian tube secretory epithelial cells. Oncogene 2016;35:2247-65. [Crossref] [PubMed]
- Lazzari C, Verlicchi A, Gkountakos A, et al. Molecular Bases for Combinatorial Treatment Strategies in Patients with KRAS mutant Lung Adenocarcinoma and Squamous Cell Lung Carcinoma. Pulm Ther 2016;2:1-18. [Crossref]
- Karachaliou N, Chaib I, Cardona AF, et al. Common Co-activation of AXL and CDCP1 in EGFR-mutation-positive Non-smallcell Lung Cancer Associated With Poor Prognosis. EBioMedicine 2018;29:112-27. [Crossref] [PubMed]
- Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. [Crossref] [PubMed]
- Balaji K, Vijayaraghavan S, Diao L, et al. AXL Inhibition Suppresses the DNA Damage Response and Sensitizes Cells to PARP Inhibition in Multiple Cancers. Mol Cancer Res 2017;15:45-58. [Crossref] [PubMed]
- Hai J, Liu S, Bufe L, et al. Synergy of WEE1 and mTOR Inhibition in Mutant KRAS-Driven Lung Cancers. Clin Cancer Res 2017;23:6993-7005. [Crossref] [PubMed]
- Uekita T, Fujii S, Miyazawa Y, et al. Oncogenic Ras/ERK Signaling Activates CDCP1 to Promote Tumor Invasion and Metastasis. Mol Cancer Res 2014;12:1449-59. [Crossref] [PubMed]
- Leroy C, Shen Q, Strande V, et al. CUB-domain-containing protein 1 overexpression in solid cancers promotes cancer cell growth by activating Src family kinases. Oncogene 2015;34:5593-8. [Crossref] [PubMed]
- Seguin L, Camargo MF, Wettersten HI, et al. Galectin-3, a Druggable Vulnerability for KRAS-Addicted Cancers. Cancer Discov 2017;7:1464-79. [Crossref] [PubMed]
- Huber KV, Salah E, Radic B, et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 2014;508:222-7. [Crossref] [PubMed]
- Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene 2009;28:3597-607. [Crossref] [PubMed]
- Justilien V, Ali SA, Jamieson L, et al. Ect2-Dependent rRNA Synthesis Is Required for KRAS-TRP53-Driven Lung Adenocarcinoma. Cancer Cell 2017;31:256-69. [Crossref] [PubMed]
- Ali SA, Justilien V, Jamieson L, et al. Protein Kinase Ciota Drives a NOTCH3-dependent Stem-like Phenotype in Mutant KRAS Lung Adenocarcinoma. Cancer Cell 2016;29:367-78. [Crossref] [PubMed]
- Justilien V, Walsh MP, Ali SA, et al. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 2014;25:139-51. [Crossref] [PubMed]
- Romero R, Sayin VI, Davidson SM, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 2017;23:1362-8. [PubMed]
- Liu H, Murphy CJ, Karreth FA, et al. Identifying and Targeting Sporadic Oncogenic Genetic Aberrations in Mouse Models of Triple-Negative Breast Cancer. Cancer Discov 2018;8:354-69. [Crossref] [PubMed]


