
Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma
Introduction
Lung cancer accounts for 17% of new cancer cases and approximately one-quarter of all cancer-related deaths globally (1). Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and is traditionally managed by surgical resection, radiotherapy, and chemotherapy. Identifying specific genetic alteration helps oncologists to stratify NSCLC patients for targeted therapy. For example, epidermal growth factor receptor (EGFR) activating mutations, including Ex19del and L858R, have been found in 10–40% of NSCLC patients, and EGFR tyrosine kinase inhibitors (EGFR-TKIs) showed promising therapeutic responses in these patients (2). Targeting other genetic alterations, such as ALK and ROS1, also helps to improve the survival of NSCLC patients (3).
Large-scale high throughput sequencing studies have revealed the complexities of the genomic landscape of NSCLC (4,5), which is associated with tumor heterogeneity and the risk of recurrence and mortality (6). Surgical or biopsy tissue samples are subject to sampling bias and sequencing these samples only gives a snapshot of tumor heterogeneity. In order to gain insights into the mutational landscapes of NSCLC and to guide targeted therapy, temporal characterization of genomic alterations of each tumor is required.
Tissue-based next-generation sequencing (NGS) is the gold-standard technique for genomic characterization; however, it requires serial biopsies and thus clinically challenging (7). In contrast, the liquid biopsy of extracellular vesicles (e.g., exosomes) and circulating tumor DNA has recently emerged as a promising non-invasive method that enables not only biomarker determination but also the temporal characterization of each tumor (8). Pleural effusion samples from NSCLC patients have been used for gene mutation analysis and its result matched with that obtained from corresponding tumor tissues (9-12).
Tumor exosome is a specific subtype of membranous microvesicle released into the tumor microenvironment (13). Recently, exosome emerged as a promising diagnostic and prognostic biomarker in cancer (8,14). miRNA isolated from circulating exosomes matched the miRNA pattern expressed in tumor tissues in NSCLC patients (15,16), and miRNAs were also found in pleural effusion-derived exosomes in NSCLC patients (17,18). Huang et al. found that 80% of the exosomes from NSCLC biopsies were EGFR positive (19). In addition, exosomal nucleic acids (exoNAs) can be used for blood-based liquid biopsy. Möhrmann et al. sequenced plasma-derived exoNAs to assess common mutation hotspots in BRAF, EGFR, and KRAS; after comparing results of plasma exoNAs with those of standard tumor FFPE samples or plasma cfDNAs in 43 patients with advanced cancers, plasma exoNAs showed higher sensitivity for detecting common BRAF, KRAS, and EGFR mutations (20). Although it is feasible to detect well-documented NSCLC-associated mutations in plasma exoNA samples, no one has conducted genetic testing on pleural effusion-derived exosomal DNA (PE-exoDNA) samples in NSCLC patients.
The current study aimed to assess the feasibility of genetic testing using PE-exoDNA. We obtained pleural effusion samples from 20 stage IV lung adenocarcinoma patients, and their PE-exoDNA and pleural effusion-derived cfDNA (PE-cfDNA) were prepared. Genetic profiling was performed using targeted NGS of 416 cancer-relevant genes in 18 PE-cfDNA and 20 PE-exoDNA samples, and the concordance of these two samples was examined, including the genomic profile of mutations in main drivers and tumor suppressors, the mutation type, and the minor allele frequency (MAF).
Methods
Patients
We retrospectively enrolled patients with histologically confirmed stage IV lung adenocarcinoma who were considered unsuitable for surgical resection of curative intent and received chemotherapy or chemotherapy plus EGFR-TKI between February 2016 and August 2017 at Zhejiang Cancer Hospital, China. All patients provided written informed consents in compliance with ethical regulations of the Zhejiang Cancer Hospital. This study was approved by the Ethics Committee of the Zhejiang Cancer Hospital (No. IRB2014-03-032). All the samples were shipped to the central laboratory of a clinical testing center (Nanjing Geneseeq Technology Inc., China) for genetic testing.
Preparation of pleural effusion supernatant fluids and exosomes
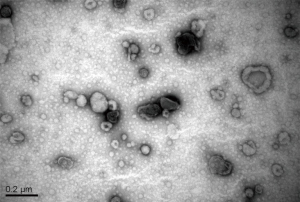
Pleural effusion was collected before initiation of EGFR-TKI therapy in a heparinized 50-mL syringe with a fine bore (18 G) needle. Samples were centrifuged at 1,800 ×g for 10 min and then 16,000 ×g for 10 min to remove cells and cell debris. The supernatants were then centrifuged at 100,000 ×g for 1 hour to collect the exosomes. The pellets were resuspended in 2-mL HEPES-saline (HBS; NaCl 150 mM, HEPES 20 mM, EGTA 2 mM, pH 7.6). Exosome quantity was estimated by Bradford dye assays (Bio-Rad). Exosomes were subjected to transmission electron microscopy for validation of exosome preparation (Figure S1). The supernatants were snap frozen and stored at −80 °C until DNA extraction.
Isolation and molecular testing of pleural effusion fluid DNA and exosomal DNA
PE-cfDNA was extracted using QIAamp Circulating Nucleic Acid Kit (Qiagen). The size distribution of cfDNA was analyzed using Bioanalyzer 2100 with the High Sensitivity DNA kit (Agilent Technologies). Exosomal DNA was extracted using the ExoLution Plus Isolation Kit (Exosome Diagnostics). All DNA samples were quantified by Qubit 3.0 using the dsDNA HS Assay Kit (Life Technologies).
Targeted NGS
The DNA library was prepared using the KAPA Hyper Prep Kit (KAPA Biosystems). Hybridization-based target enrichment was carried out with GeneseeqOne pan-cancer gene panel (416 cancer-relevant genes, Figure S2) (Nanjing Geneseeq Technology Inc., China) and xGen Lockdown Hybridization and Wash Reagents Kit (Integrated DNA Technologies). Captured libraries by Dynabeads M-270 (Life Technologies) were amplified in KAPA HiFi HotStart ReadyMix (KAPA Biosystems) and quantified by qPCR using the KAPA Library Quantification Kit (KAPA Biosystems) for sequencing.
The libraries were paired-end sequenced on the Illumina HiSeq4000 NGS platforms (Illumina). Trimmomatic (21) was used for FASTQ file quality control (QC). Leading/trailing low quality (quality reading below 15) or N bases were removed. Qualified reads were aligned to the hg19 genome using Burrows-Wheeler Aligner (BWA-mem, v0.7.12; https://github.com/lh3/bwa/tree/master/bwakit). Single nucleotide variant and indel calling were performed using VarScan2 (22). Common single-nucleotide polymorphisms (SNPs) were removed if they were present in >1% population frequency in the 1,000 Genomes Project or 65,000 exomes project (ExAC) Database, followed by annotation using ANNOVAR (23). ADTEx (http://adtex.sourceforge.net) was used to identify copy number variations (CNVs) using a normal human HapMap DNA sample NA18535. Genomic fusions were identified by FACTERA (24) with the default setting. Called-out variants were further filtered by public databases containing germline mutations, including dbSNP, 1000G, and ExAC, and any variations presented in these databases were removed.
Results
Patient characteristics and overall mutation profiles
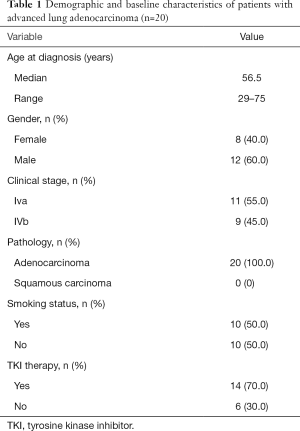
Demographic and baseline characteristics of the 20 patients in this study are summarized in Table 1. All 20 patients had stage IV lung adenocarcinoma (11 IVa and 9 IVb). Their median age was 56.5 years (range: 29–75 years) and more than half of the patients were male. Half of them had smoking history and 14 patients (70%) had received TKI therapies. Targeted NGS was conducted using PE-exoDNA samples from all 20 patients and PE-cfDNA samples from 18 patients. A total of 304 genetic alterations in 161 genes (mean 16.9 variations per patient; range: 6–41) were detected in the 18 PE-cfDNA samples, and 276 genetic changes in 152 genes (mean 13.8 variations per patient; range: 6–24) were detected in the 20 PE-exoDNA samples. For the 18 patients with both PE-exoDNA and PE-cfDNA, we detected a total of 251 variants in 152 genes (mean 13.9 variations per patient; range: 6–24) from PE-exoDNA samples.
Full table
Comparable genetic profiles between PE-exoDNA and PE-cfDNA
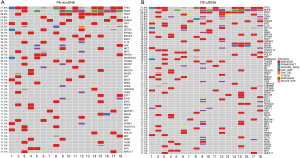
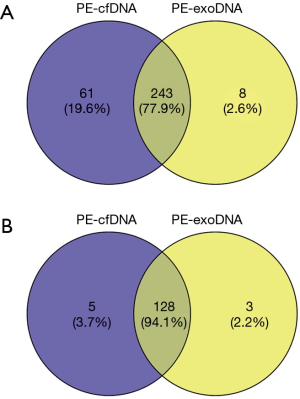
For the 18 patients with both PE-exoDNA and PE-cfDNA samples, altered genes that were detected in at least 2 patients are shown in Figure 1, and the mutation patterns were similar between the two types of DNA samples. Forty-seven mutations were detected in PE-exoDNA samples (Figure 1A) and 66 mutations were found in PE-cfDNA samples (Figure 1B). Within these identified mutated genes, 34 genes had exactly the same mutation rate when comparing PE-exoDNA and PE-cfDNA samples (Figure 1). Based on PE-cfDNA results, TP53, EGFR, PKD1, ALK, DDR2, RB1, FANCA, FAT1, FLT4, and SETD2 were top altered genes with mutation frequency >20%; similar frequently mutated genes (>20%) were detected in PE-exoDNA, including TP53, EGFR, PKD1, ALK, FANCA, FAT1, RB1, and SETD2. A total of 312 gene mutations were detected when combining the results of PE-exoDNA and PE-cfDNA, and 243 (77.9%) of them were shared by both samples; 61 gene alterations were found only in PE-cfDNA samples and 8 gene alterations were found only in PE-exoDNA samples (Figure 2A). Strikingly, most of the exclusive genetic changes were CNVs, and if we excluded CNVs from the analysis, the concordance rate between the 2 samples increased to 94.1% (Figure 2B). These results suggest that the general genetic profiles of PE-exoDNA and PE-cfDNA were highly concordant.
High concordance of mutations in driver genes and tumor suppressors between PE-cfDNA and PE-exoDNA
We then examined the distribution of specific driver genes and tumor suppressors with PE-exoDNA vs. PE-cfDNA (Figure 3). Eight genes were investigated, including EGFR, ALK, KRAS, BRAF, MET, PTEN, TP53, and RB1. In the 18 patients with both PE-exoDNA and PE-cfDNA samples, a total of 46 mutations within these 8 genes were identified. For the 2 patients with only PE-exoDNA samples, ALK fusion and TP53 in-frame shift were identified. In the 18 patients with both types of samples, 43 mutations were detected in PE-exoDNA and 45 mutations were detected in PE-cfDNA. One (1/43, 2.3%) mutation (MET amplification) was only detected in PE-exoDNA; 3 (3/45, 6.7%) mutations (MET amplification, EGFR amplification, and RB1 single copy loss) were found only in PE-cfDNA. Seven patients harbored EGFR Ex19del, 2 harbored the L858R, and 1 had both Ex19del and L858R. In the patients with EGFR activating mutations, 2 harbored MET amplification and 6 harbored EGFR amplification. The remaining cases had ALK fusion (2, 11.1%), KRAS activating mutations (2, 11.1%), BRAF V600E (1, 5.6%), and PTEN splice-site mutation (1, 5.6%). Taken together, 42 mutations were identified in both PE-exoDNA and PE-cfDNA, with a concordance of 91.3%. This finding suggests that PE-exoDNA could be used for mutation testing of driver genes and tumor suppressors in stage IV lung adenocarcinoma patients.

High concordance of mutation type between PE-exoDNA and PE-cfDNA
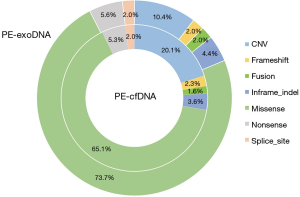
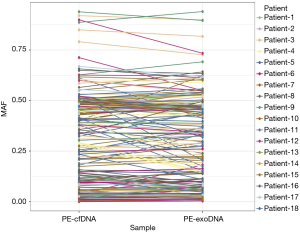
Missense mutations accounted for 73.7% and 65.1% of all mutations in PE-exoDNA and PE-cfDNA, respectively (Figure 4). PE-cfDNA and PE-exoDNA had comparable percentage of frameshift (2.3% vs. 2.0%), gene fusion (1.6% vs. 2.0%), in-frame-indel (3.6% vs. 4.4%), nonsense mutation (5.3% vs. 5.6%), and splice-site mutation (2.0% vs. 2.0%). PE-cfDNA had a higher percentage of CNV than PE-exoDNA (20.1% vs. 10.4%), suggesting that targeted NGS may be superior in detecting CNV in PE-cfDNA vs. PE-exoDNA. Finally, we compared the mutation MAF between PE-cfDNA and PE-exoDNA. As shown in Figure 5, the paired MAFs for mutations in each patient were likely to be similar for the two samples as there was no statistically significant difference (Wilcoxon test, P=0.068), implying that the results of PE-exoDNA and PE-cfDNA were generally comparable.
Treatment
In the 18 patients with both PE-exoDNA and PE-cfDNA samples, 7 patients harbored EGFR Ex19del, 2 harbored the L858R, 1 had both Ex19del and L858R, and 2 had ALK fusion. The concordance rate of EGFR mutations and ALK fusions was 100% in the 18 patient samples. Among the 10 patients with EGFR mutations, all received EGFR-TKIs (7 with icotinib and 3 with gefitinib). The median progression-free survival was 10.6 months and the objective response rate was 70%. For the 2 patients harboring ALK fusions, the median progression-free survival was 12.0 months with crizotinib treatment.
Discussion
There has been a growing interest in exosomes as a source of cancer biomarkers. Recently, human body fluid-derived exoNA has been used to perform genetic testing. Möhrmann et al. used NGS to compare common mutation hotspots in BRAF, EGFR, and KRAS detected in plasma exoNA versus tumor FFPE exoNA or plasma cfDNA in patients with advanced cancers (20). Of the 41 mutations detected in tumor tissues, 39 were detected in plasma exoNA, with an overall sensitivity of 95% (95% CI, 83–99%), suggesting that plasma-derived exoNA could be used for genetic testing in advanced cancers. However, it was still unknown whether targeted NGS using exosomal DNA from malignant pleural effusion was feasible. In the current study, we presented the first piece of convincing evidence that malignant PE-exoDNA and PE-cfDNA exhibited highly concordant mutational profiles.
Oncoprint can provide a concise graphical summary of genomic alterations in multiple genes across a set of tumor samples (25,26). In the current study, we generated a profile that consisted of 46 genetic alterations in 8 genes by comparing PE-cfDNA and PE-exoDNA samples in 18 patients with advanced lung adenocarcinoma. Forty-three abnormalities were identified in PE-exoDNA; 45 were identified in PE-cfDNA; 42 were shared between PE-cfDNA and PE-exoDNA. The concordance rate was 91.3% between the 2 types of samples. When CNVs were excluded from the analysis, the concordance rate between PE-cfDNA and PE-exoDNA samples was 94.1%. To our knowledge, this is the first study that systematically assessed the concordance of genomic alterations across a large number of genes in both PE-cfDNA and PE-exoDNA samples. Our findings demonstrate that PE-exoDNA could yield a similar mutation profile compared with that of PE-cfDNA, especially in driver genes and some important tumor suppressors.
Although the genetic profile of cfDNA and exoDNA had a very high concordance in our lung cancer patient cohort, exoDNA potentially has more clinical utility than cfDNA. Compared with cfDNA that is solely diluted in plasma, pleural effusion, or other body fluids, exoDNA is contained in exosomes that could also carry cancer-related proteins and RNAs. Multiple studies have demonstrated that these exosomal proteins and RNAs could serve as biomarkers (27), which can be used to cross-validate the exoDNA results and provide more comprehensive clinical information for cancer diagnosis and treatment. Furthermore, one challenge to use these extracellular DNAs is to distinguish cancer-derived cfDNAs/exoDNAs from those derived from normal cells. It is not easy to globally enrich tumor-derived cfNDA. In contrast, each exosome exposes 10–100 surface antigens, and exosomal surface EGFR and LRG1 have been identified in lung cancer patients (28,29). These cancer-specific surface markers can be used to isolate tumor exosomes, and thus enriching tumor exoDNAs. These potential clinical advantages of exoDNAs need to be validated in future studies.
The current study has several limitations. First, the sample size was relatively small, with only 20 patients with lung adenocarcinoma. Also, PE-cfDNA was available for only 18 patients. Second, we did not compare the results obtained from PE-exoDNA or PE-cfDNA with tumor tissue samples. Future studies with larger sample size and corresponding tumor tissues are needed to verify our results.
In conclusion, targeted NGS analysis demonstrated similar genetic profile obtained from PE-exoDNA versus PE-cfDNA in patients with advanced lung adenocarcinoma. These findings encourage future exploration of exoDNA as an alternative biological source for clinical diagnosis for lung cancer.
Acknowledgments
Funding: The study was funded by grants from the Medical Scientific Research Foundation of Zhejiang Province (No. 2015KYA040 and 2016KYB046) and National Natural Science Foundation of China (grant 81802276).
Footnote
Conflicts of Interest: J Yan is the employee of Nanjing Geneseeq Technology Inc.; YW Shao is the employee of Geneseeq Technology Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients provided written informed consents in compliance with ethical regulations of the Zhejiang Cancer Hospital. This study was approved by the Ethics Committee of the Zhejiang Cancer Hospital (No. IRB2014-03-032).
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn 2013;15:415-53. [Crossref] [PubMed]
- Hanna N, Johnson D, Temin S, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2017;35:3484-515. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Ilié M, Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res 2016;5:420-3. [Crossref] [PubMed]
- Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, et al. Exosomes - structure, biogenesis and biological role in non-small-cell lung cancer. Scand J Immunol 2015;81:2-10. [Crossref] [PubMed]
- Kimura H, Fujiwara Y, Sone T, et al. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer 2006;95:1390-5. [Crossref] [PubMed]
- Soh J, Toyooka S, Aoe K, et al. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int J Cancer 2006;119:2353-8. [Crossref] [PubMed]
- Liu D, Lu Y, Hu Z, et al. Malignant pleural effusion supernatants are substitutes for metastatic pleural tumor tissues in EGFR mutation test in patients with advanced lung adenocarcinoma. PLoS One 2014;9:e89946. [Crossref] [PubMed]
- Guan Y, Wang ZJ, Wang LQ, et al. Comparison of EGFR mutation rates in lung adenocarcinoma tissue and pleural effusion samples. Genet Mol Res 2016;15. [Crossref] [PubMed]
- Andre F, Schartz NE, Movassagh M, et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002;360:295-305. [Crossref] [PubMed]
- Huang C, Liu S, Tong X, et al. Extracellular vesicles and ctDNA in lung cancer: biomarker sources and therapeutic applications. Cancer Chemother Pharmacol 2018;82:171-83. [Crossref] [PubMed]
- Rabinowits G, Gercel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. [Crossref] [PubMed]
- Giallombardo M, Chacartegui Borras J, Castiglia M, et al. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients' Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J Vis Exp 2016. [Crossref] [PubMed]
- Rodríguez M, Silva J, Lopez-Alfonso A, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer 2014;53:713-24. [PubMed]
- Wang Y, Xu YM, Zou YQ, et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine (Baltimore) 2017;96:e8361. [Crossref] [PubMed]
- Huang SH, Li Y, Zhang J, et al. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest 2013;31:330-5. [Crossref] [PubMed]
- Möhrmann L, Huang HJ, Hong DS, et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin Cancer Res 2018;24:181-8. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med 2013;7:769-78. [Crossref] [PubMed]
- Li Y, Zhang Y, Qiu F, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 2011;32:1976-83. [Crossref] [PubMed]
- Yamashita T, Kamada H, Kanasaki S, et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013;68:969-73. [PubMed]


