
Emerging oncogenic fusions other than ALK, ROS1, RET, and NTRK in NSCLC and the role of fusions as resistance mechanisms to targeted therapy
Introduction
Oncogenic driver mutations are known to play important roles in carcinogenesis and tumor progression in some non-small cell lung cancers (NSCLCs), especially in lung adenocarcinomas. Epidermal growth factor receptor (EGFR) mutations or anaplastic lymphoma kinase (ALK) fusions represent such oncogenic driver mutations in NSCLC, and pharmacologic inhibition using specific tyrosine kinase inhibitors (TKIs) in NSCLC patients harboring these oncogenic driver mutations has revolutionized treatment in advanced stage diseases (1). In addition to these two “classical” oncogenic driver mutations, ROS1 fusion and BRAF V600E mutation are predictive biomarkers already approved for clinical use, and neurotrophic receptor tyrosine kinase (NTRK) fusions, MET exon 14 skipping mutations, ERBB2 exon 20 insertion mutations, RET fusions, and KRAS G12C mutation have joined the list of treatable oncogenic driver mutations in NSCLCs (2-7).
Recent advances in sequencing technology have shown that gene fusion, caused by chromosomal rearrangements, is one of the frequent hallmarks of cancer genome aberrations. For example, a detailed analysis of The Cancer Genome Atlas (TCGA) dataset identified 20,731 gene fusions in 9,966 well-characterized cancer samples across 33 cancer types (after filtering against a list of 3,838 transcript “fusions” detected in a panel of 648 non-neoplastic samples) (8). Another study that analyzed 9,624 tumors from TCGA identified a total of 25,664 fusions and suggested that fusions drive the development of 16.5% of cancer cases and function as the sole oncogenic driver in more than 1% of cancer cases (9). In this review, we will summarize and discuss novel gene fusions, other than ALK, ROS1, NTRK, and RET fusions, that are considered to be oncogenic drivers in NSCLCs, especially for lung adenocarcinomas. These rare but potentially important fusions include neuregulin-1 (NRG1) fusions, MET fusions, fusion genes involving fibroblast growth factor receptor (FGFR) family members, EGFR fusions, and BRAF fusions. Some studies reported that rare primary pulmonary tumors have specific fusion genes, e.g., synaptotagmin 1 (SYT)-SSX1 or SYT-SSX2 fusions in synovial sarcoma (10) and EWS RNA binding protein 1 (EWSR1)-cAMP responsive element binding protein 1 (CREB1) fusion in pulmonary myxoid sarcoma (11); however, we will not include these rare tumors in this review.
Mutational processes of gene fusions in lung adenocarcinomas
In considering the mutational processes of lung adenocarcinomas, it is important to classify lung adenocarcinomas into two groups: lung adenocarcinomas unrelated to smoking and those related to smoking (12). A recent study by Lee et al. (13) used a mutational signature 4 (a C:G>>A:T-dominant signature, related to exposure to smoking carcinogen) (14) as a negative marker for lung adenocarcinomas unrelated to smoking. In their analyses, lung adenocarcinomas unrelated to smoking were further classified into two groups: tumors with oncogene mutations (EGFR, KRAS G12D or G12A, ERBB2, and MET exon 14 skipping) and those with oncogenic gene fusions (ALK, ROS1, RET, fibroblast growth factor receptor 2 (FGFR2), neuregulin-1 (NRG1), MET, and AXL). In the analysis of tumors with oncogenic gene fusions, the authors found that 26% of oncogenic fusion genes were generated by simple rearrangements such as large deletions (e.g., EZR-ROS1), reciprocal inversions (e.g., EML4-ALK and KIF5B-RET), and reciprocal translocations (e.g., CD74-ROS1). In contrast, 74% of oncogenic gene fusions, including all NRG1, AXL, FGFR2, and MET fusions, were complex and involved a median of 20 rearrangement breakpoints (range, 4–281). These complex rearrangements were considered to be generated by chromoplexy (15) or chromothripsis, a mutational process involving catastrophic chromosomal shattering followed by stochastic rejoining of the DNA segments (16). In addition, careful reconstruction of the complex rearrangements provided evidence of secondary complex rearrangements in some cases superimposed on the oncogenic fusion gene-generating chromoplexy. These results indicate that there are numerous possibilities for oncogenic fusion genes in lung adenocarcinomas unrelated to smoking. In the following sections, we mainly focus on the recurrent oncogenic gene fusions in lung adenocarcinomas that can be targetable using molecular targeted drugs.
NRG1 fusions
NRG1 is a ligand for ERBB3 and ERBB4 receptor tyrosine kinases (17) that is proteolytically cleaved and secreted. NRG1 is involved in a diverse spectrum of cellular processes primarily in, but not limited to, neural and cardiac development. The possible occurrence of the NRG1 fusion in lung cancer was described in 2004 (18), the same year when activating EGFR mutations were discovered in NSCLCs (19,20). The authors focused on a recurrent chromosome breakpoint in breast cancer at the NRG1 gene, and the study included 11 NSCLC specimens, with one positive case (with squamous cell histology).
A decade later, Fernandez-Cuesta and colleagues discovered a novel chimeric transcript that fused CD74 to the EGF-like domain of the NRG1 III-beta3 isoform in lung adenocarcinoma cases with invasive mucinous subtype (21). Mechanistically, part of CD74 replaced the transmembrane domain of wild-type NRG1 III-beta3 but preserved the membrane-tethered extracellular EGF-like domain of NRG1 III-beta3, thereby providing a ligand for ERBB3/ERBB2 receptor complexes. Mechanistically, it is considered that binding of the EGF-like domain of the NRG1 fusion to ERBB3 in an autocrine, paracrine, or juxtacrine fashion triggers the activation of ERBB2/ERBB3 complex and the downstream signaling (Figure 1) (22). Therefore, it is reasonable that the authors observed that ERBB2 and ERBB3 expression was high along with high phosphorylated ERBB3 levels in tumors bearing the CD74-NRG1 fusion (21). After this initial study, three additional groups reported the presence of NRG1 fusions in NSCLCs in 2014, expanding NRG1 fusion partners to solute carrier family 3 member 2 (SLC3A2), syndecan 4 (SDC4), and others (23-25). To date, while many genes have been reported as the partners of NRG1 fusions (26), CD74-NRG1 accounts for approximately half of cases in NSCLCs with NRG1 fusions (26,27).
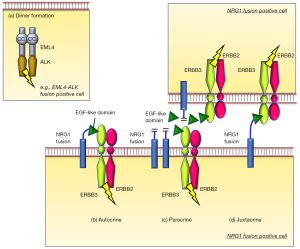
Intensive analyses showed that the incidence of NRG1 fusions in NSCLC is very rare. A recent large-scale study by Jonna and colleagues evaluated the incidence of NRG1 fusions in 21,858 solid tumor specimens profiled at a genomics laboratory, Caris Life Sciences, from September 2015 to December 2018. The authors found that only 0.3% of NSCLCs, predominantly tumors that contain invasive mucinous part (32%), had NRG1 fusions (27). The incidence of NRG1 fusion was recently reported as even lower in Chinese patients (0.16%, 18 of 10,966 NSCLC patients) (28), while another recent report from China found an incidence similar with rates in Caucasian populations (0.36%, 6 of 1,681 lung adenocarcinomas) (29). The study by Jonna and colleagues (27) also reported that NRG1 fusions could be detected at a low incidence across multiple tumor types, e.g., 0.5% of gallbladder cancers, 0.5% of pancreatic ductal adenocarcinomas, 0.4% of ovarian cancers, and 0.2% of breast cancers; however, CD74 was not detected as the partner gene in tumors other than NSCLCs.
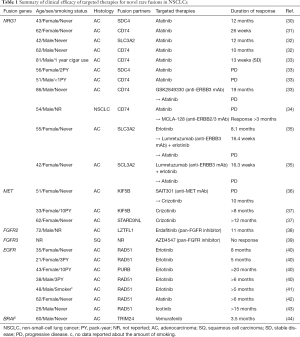
Because NRG1 fusions are considered to bind ERBB3, and ERBB3/ERBB2 heterodimers activate downstream signaling, molecular targeted drugs that inhibit this pathway are anticipated to show efficacy in tumors with this fusion. The first attempt, as case studies, was performed in 2017 (30-32) by three independent groups using afatinib, an irreversible pan-ERBB TKI (EGFR, ERBB2, and ERBB4), which has been approved in the treatment of NSCLCs with activating EGFR mutations (1). Four NRG1 fusion positive lung adenocarcinoma patients (two with CD74-NRG1, one with SLC3A2-NRG1, and one with SDC4-NRG1) received afatinib monotherapy, which led to clinical benefit lasting from 26 weeks to 12 months (Table 1). Other studies reported that GSK2849330, an anti-ERBB3 monoclonal antibody (mAb) (33), MCLA-128 (zenocutuzumab), a bispecific ERBB2/3 antibody (34), and the combination of lumretuzumab (anti-ERBB3 mAb) plus erlotinib (35) showed clinical efficacy in one or a few NSCLC patients with NRG1 fusions. It should be noted that, in contrast to aforementioned case reports of afatinib, some of these patients experienced progressive disease following afatinib monotherapy (Table 1). Notably, an in vitro study using afatinib, pertuzumab (anti-ERBB2 mAb), and lumretuzumab against an SLC3A2-NRG1 fusion model reported that the combination treatment with two mAbs or the combination treatment of one of the drugs plus taxol was more effective than each of the single agents alone (45). These in vitro studies will be important to evaluate the effectiveness of potential treatment strategies, although the results should be confirmed in clinical settings.
Full table
MET fusions
MET gene aberration by exon 14 skipping has become an important therapeutic target in lung adenocarcinomas and possibly in pleomorphic carcinomas of the lung (4). In addition, several studies reported MET activation in lung adenocarcinomas by MET gene fusions.
MET gene fusions with KIF5B is a recurrent fusion, although the incidence is quite low, as reported by some independent research groups (36,37,46,47). A recent study by Gow and colleagues reported the oncogenic activity of KIF5B-MET by soft agar colony formation assays and a xenograft mouse model, and the authors found that crizotinib effectively inhibited the growth of tumors harboring these fusions in vitro and in vivo (47). Notably, two patients with KIF5B-MET fusion and one patient with STARD3NL-MET fusion received crizotinib (Table 1) and all three patients showed clinical benefit (36,37). Recently, additional novel fusions involving MET, CD47-MET (29), HLA-DRB1-MET (48) and MET-ATXN7L1 (49) were also reported in a common-driver negative lung adenocarcinoma patient.
FGFR fusions
Gene fusions involving FGFR family members were discovered in glioblastoma multiforme in 2012, such as FGFR1 and FGFR3 fusions with transforming acidic coiled-coil containing protein 1 (TACC1) and TACC3, respectively (50), and in bladder carcinomas in 2013 (FGFR3-TACC3 fusion) (51). Subsequent analysis across multiple tumor cohorts (52) revealed that FGFR fusions are present in a wide variety of tumors including lung squamous cell carcinomas, as summarized below. Although the FGFR3-TACC3 fusion is more common in lung squamous cell carcinomas, this FGFR3-TACC3 fusion has also been identified in lung adenocarcinomas (38,53,54). In addition, a recent report observed the presence of FGFR1 fusions (BAG4-FGFR1, which has been previously reported in lung squamous cell carcinomas (54), and FGFR1-CIT), FGFR2 fusions (FGFR2-KIAA1598, which was previously described in cholangiocarcinoma (55), FGFR2-CIT, FGFR2-ERC1, FGFR2-LZTFL1, FGFR2-POC1B, FGFR2-SORBS1, FGFR2-TP73, FGFR2-TXLNA), FGFR3 fusions other than FGFR3-TACC3 (FGFR3-PHLDB3 and WHSC1-FGFR3), and FGFR4 fusions (ANO3-FGFR4 and NSD1-FGFR4) in lung adenocarcinomas, adenosquamous cell carcinomas, or NSCLC not otherwise specified (38). Among patients with these FGFR fusions, one invasive mucinous adenocarcinoma patient with FGFR2-LTZTFL1 fusion was treated with erdafitinib, a pan-FGFR inhibitor, in a clinical trial; he attained a partial response with 60% tumor shrinkage after 2 months of therapy and continued to receive this drug for a total of 11 months (38) (Table 1).
Other rare fusions in lung cancers
Through detailed analyses, such as through RNA-based next-generation sequencing and fusion assay, of selected patients with tumors without known driver mutations (e.g., EGFR, KRAS, ERBB2, BRAF V600E, ALK, ROS1) or tumors with invasive mucinous phenotype (enriched cohort for NRG1 fusions), several rare fusions have been identified. Nakaoku and colleagues conducted whole-transcriptome sequencing for 32 invasive mucinous adenocarcinoma tissues without KRAS mutation and detected one EZR-ERBB4 fusion and one TRIM24-BRAF fusion in addition to six NRG1 fusions and one novel RET fusion (KIAA1468-RET) (23). The tumorigenicity of NIH-3T3 cells expressing EZR-ERBB4 or TRIM24-BRAF fusion cDNAs was confirmed through experiments using nude mine. A 60-year-old male lung adenocarcinoma patient with TRIM24-BRAF fusion was reported to respond to vemurafenib; however, the duration of response was only 3.5 months (44) (Table 1). A recently reported technique involving a single-tube, dual-template assay and an integrated bioinformatics pipeline for relevant variant calling identified the BBS9-BRAF fusion in lung adenocarcinomas (48). Another study reported five SND1-BRAF fusions in lung adenocarcinoma tissues; however, the role of SND1-BRAF fusion as an oncogenic driver is unclear, since four out of five detected fusions co-existed with other driver mutations (two EGFR exon 20 mutations, one ERBB2 YVMA insertion, and one EML4-ALK fusion) (56). The SND1-BRAF fusion gene was also reported as a potential resistance mechanism to a MET inhibitor, PF-04217903, in GTL16 gastric adenocarcinoma cells with MET gene amplification through MAPK activation (57).
Activation of EGFR by a recurrent gene fusion (EGFR-RAD51 fusion) was reported in 2016 in 4 out of ~10,000 lung adenocarcinomas (40). Tumors bearing this EGFR-RAD51 fusion, as well as those with EGFR-PURB fusion, were markedly sensitive to EGFR-TKIs (40), which was confirmed by recent case studies (41-43) (Table 1). Recent reports identified several rare fusion genes for EGFR, such as SEPT14-EGFR, EGFR-KDD, EGFR-YAP1, EGFR-SHC1, and others, in NSCLCs at frequencies of 0.0–0.13% (29,42,43). Another group reported the presence of EGFR-ANXA2 and EGFR-RAD51 double fusion mutations in a 36-year-old female patient with lung adenocarcinoma (58).
Fusion genes in lung squamous cell carcinomas
Fusion genes are also frequently found in lung squamous cell carcinomas (59); however, their potential as therapeutic targets are largely unknown. One exception is the FGFR3-TACC3 fusion, which was identified by independent research groups in 0.6–1.3% of lung squamous cell carcinomas (9,38,52,54,60,61). Interestingly, the FGFR3-TACC3 fusion is also found in head and neck squamous cell carcinomas, oral cancers, cervical squamous cell carcinomas, and bladder cancers (9,52) as well as lung adenocarcinomas, as described above (38). In vivo experiments of mice harboring tumors derived from RT4 and SW780 bladder cancer cells, containing FGFR3-TACC3 and FGFR3-BAIAP2L1 fusion, respectively, showed that a FGFR inhibitor, PD173074, inhibited tumor growth in a dose-dependent manner (52). However, in a phase II study of a FGFR inhibitor, AZD4547, in previously treated patients with lung squamous cell carcinoma with FGFR aberration(s), only one patient had FGFR3 fusion among those included in this study, and the patient did not respond to this novel agent (39) (Table 1). Several phase I studies of pan-FGFR inhibitors have been performed in solid tumors with genetic aberration(s) of FGFRs (gene amplifications, mutations, and fusions); however, the response rates of lung squamous cell carcinomas or NSCLCs were as low as 5% compared with 46% and 27% in urothelial carcinoma and cholangiocarcinomas, respectively (62). Other fusion genes as possible oncogenic drivers in lung squamous cell carcinomas include BAG4-FGFR1 fusion (52,54) and FGFR2-KIAA1967 fusion (52). The above-mentioned NRG1 fusions were also detected in two lung squamous cell carcinomas out of 9,592 NSCLCs (27).
Fusions as a mechanism of acquired resistance to TKIs
The first choice of treatment for advanced NSCLCs with driver mutations, such as EGFR mutations, ALK fusions, or ROS1 fusions, is TKI monotherapy that targets specific molecules (1). However, despite initial dramatic clinical responses, the emergence of acquired resistance to these molecular targeted therapies is almost inevitable (63). Analyses of the molecular mechanisms underlying the acquired resistance to TKIs have been extensively performed in NSCLCs. These resistance mechanisms can be classified into several groups including (I) a secondary mutation/amplification of the targeted molecule, such as T790M secondary mutation in EGFR, (II) activation of a bypass pathway, such as MET gene amplification, (III) activation of a downstream pathway, such as PTEN loss, in tumors with EGFR mutation, and (IV) histological/morphological transformation including small cell lung cancer transformation and epithelial to mesenchymal transition (63).
The increasing use of comprehensive genomic testing and wide application of re-biopsy (including liquid biopsy) at the time of resistance has expanded our understanding of TKI resistance mechanisms in NSCLCs. Despite initial belief that oncogenic fusion genes such as ALK fusion (64) or ROS1 fusion (65) are mutually exclusive with other oncogenic driver mutations in NSCLCs, since 2015 (66), many reports have suggested that gene fusion sometimes causes acquisition of resistance to TKI monotherapy through the activation of bypass pathways (67).
How, then, do fusion genes occur in lung adenocarcinoma patients with EGFR mutation? The aforementioned study by Lee et al. that traced gene fusions in the mutational history of lung adenocarcinomas has provided a possible answer for this question (13). As described above, the authors classified lung adenocarcinomas unrelated to smoking [low S4 signature (14)] into tumors with oncogenic mutations and those with oncogenic gene fusions. Interestingly, the authors observed a significantly larger burden of rearrangements in tumors with oncogenic mutations than in those with oncogenic gene fusions (211 versus 87; P=0.0009). EGFR activating mutation, as well as other oncogenic mutations, occurs in the early stage of carcinogenesis, and these oncogenic mutations are suggested to accelerate the occurrence of chromosomal rearrangements in the later stage of tumor development. Some rearrangements accidentally involve oncogenes such as RET or ALK, and minor clones with RET or ALK fusion will be dominant upon EGFR-TKI treatment.
The roles of receptor tyrosine kinase (RTK) fusions as a resistance mechanism to EGFR-TKIs in NSCLCs with EGFR mutation have been comprehensively summarized in a very recent review by Zhu and colleagues (67). In this review, the authors identified a total of 86 cases with RTK gene fusion that acquired resistance to EGFR-TKI(s). Acquired RTK gene fusions were observed at progression across all three generations of EGFR-TKIs, but were apparently enriched after osimertinib therapy (3.7%, 15/409 cases) compared with after 1st or 2nd generation EGFR-TKI therapy (1.8%, 3/167 cases). This difference would be reasonable, since more than half of patients who progress against 1st or 2nd generation EGFR-TKIs harbor a T790M secondary mutation as a resistance mechanism (68).
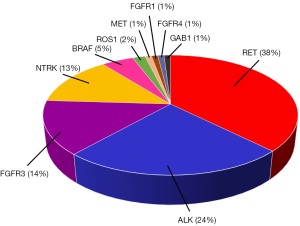
In addition to the 86 cases summarized in the above review (67), our literature search identified an additional thirteen patients who acquired resistance to EGFR-TKIs and harbored gene fusions (38,69-72), including BRAF fusions. We could not find NRG1 fusion as the mechanism of acquired resistance to EGFR-TKIs. The rates of fusion genes as a resistance mechanism to EGFR-TKIs are summarized in Figure 2, with the highest incidence in RET gene fusions, followed by ALK, FGFR3, and NTRK fusions. As reported in the review by Zhu and colleagues (67), interestingly, KIF5B accounts for only 2% of the fusion partners of acquired resistance RET fusions, whereas it accounts for 54% in a survey of 106 Chinese NSCLC patients with RET fusions. In contrast, CCDC6-RET accounts for 17% of the de novo RET fusions in NSCLCs compared with 58% of the RET fusions related to EGFR-TKI resistance. Although the data are limited, safety and clinical efficacy of a dual blockade of acquired gene fusions and mutated EGFR (founder) were reported in 10 cases (67).
Acquired gene fusion as a resistance mechanism to TKIs may occur in patients with a founder ALK fusion who received ALK-TKIs. Through comprehensive analyses for resistance mechanisms to ALK-TKIs in 43 patients with founder ALK fusions, McCoach and colleagues identified a RALGAPA1-NRG1 fusion in a post-alectinib tumor sample and a CCDC6-RET fusion in a post-brigatinib biopsy tissue that was not detected on the pre-brigatinib biopsy performed after alectinib treatment. In vitro analysis showed that CRISPR-induced RALGAPA1-NRG1 fusion conferred crizotinib resistance in the ALK-positive lung cancer cell line, H3122, and addition of afatinib re-sensitized H3122 cells with RALGAPA1-NRG1 fusion to crizotinib (73). The authors also evaluated 12 patients with founder ROS1 fusions; however, no acquired gene fusion was detected (73). Recently, Boyle and colleagues reported an AGK-BRAF fusion as a potential resistance mechanism to ALK inhibitor therapy in a lung adenocarcinoma patient with founder EML4-ALK fusion (74).
Conclusions
The increasing use of comprehensive genomic profiling in research as well as the clinical setting has contributed to accumulation of numerous data regarding gene fusions. However, we should recognize that some comprehensive panel tests analyze both DNA and RNA samples while others examine DNA only, and that the latter may not be able to detect rare novel fusions often due to the large intronic regions. We now know that gene rearrangement is a frequent event in the initiation and progression of NSCLCs. Several novel fusions, in addition to ALK, ROS1, RET, and NTRK fusions, are considered to be oncogenic drivers in NSCLCs. In addition, evidence has suggested that acquisition of gene fusion is a molecular mechanism of acquired resistance to targeted therapies. Detection, validation, and pharmacological inhibition of these fusion genes are becoming more important in the treatment of NSCLC patients.
Acknowledgments
We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Funding: This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (18K07336 to K Suda) and by a grant from the Uehara Memorial Foundation to K Suda.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Silvia Novello, Francesco Passiglia) for the series “Looking for Chimeras in NSCLC: Widen Therapeutic Options Targeting Oncogenic Fusions” published in Translational Lung Cancer Research. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-186). The series “Looking for Chimeras in NSCLC: Widen Therapeutic Options Targeting Oncogenic Fusions” was commissioned by the editorial office without any funding or sponsorship. TM serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. KS reports grants and personal fees from Boehringer Ingelheim, personal fees from AstraZeneca, grants from Rain Therapeutics, outside the submitted work. TM reports grants and personal fees from Boehringer Ingelheim, personal fees from AstraZeneca, grants and personal fees from Pfizer Japan Inc., grants and personal fees from Chugai Pharmaceutical Co. Ltd., personal fees from MSD K. K., grants and personal fees from Ono Pharmaceutical Co. Ltd., personal fees from Bristol Myers Squibb, personal fees from Eli Lilly Japan K. K., grants and personal fees from Taiho Pharmaceutical Co. Ltd., outside the submitted work. Both authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hirsch FR, Suda K, Wiens J, et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388:1012-24. [Crossref] [PubMed]
- Larotrectinib OK'd for Cancers with TRK Fusions. Cancer Discov 2019;9:8-9.
- Entrectinib OK'd for Cancers with NTRK Fusions, NSCLC. Cancer Discov 2019;9:OF2. [Crossref]
- Fujino T, Kobayashi Y, Suda K, et al. Sensitivity and Resistance of MET Exon 14 Mutations in Lung Cancer to Eight MET Tyrosine Kinase Inhibitors In Vitro. J Thorac Oncol 2019;14:1753-65. [Crossref] [PubMed]
- Koga T, Kobayashi Y, Tomizawa K, et al. Activity of a novel HER2 inhibitor, poziotinib, for HER2 exon 20 mutations in lung cancer and mechanism of acquired resistance: An in vitro study. Lung Cancer 2018;126:72-9. [Crossref] [PubMed]
- Guo R, Schreyer M, Chang JC, et al. Response to Selective RET Inhibition With LOXO-292 in a Patient With RET Fusion-Positive Lung Cancer With Leptomeningeal Metastases. JCO Precis Oncol 2019. [Crossref] [PubMed]
- Hallin J, Engstrom LD, Hargis L, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10:54-71. [Crossref] [PubMed]
- Hu X, Wang Q, Tang M, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 2018;46:D1144-9. [Crossref] [PubMed]
- Gao Q, Liang WW, Foltz SM, et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep 2018;23:227-38.e3. [Crossref] [PubMed]
- Yano M, Toyooka S, Tsukuda K, et al. SYT-SSX fusion genes in synovial sarcoma of the thorax. Lung Cancer 2004;44:391-7. [Crossref] [PubMed]
- Matsukuma S, Hisaoka M, Obara K, et al. Primary pulmonary myxoid sarcoma with EWSR1-CREB1 fusion, resembling extraskeletal myxoid chondrosarcoma: Case report with a review of Literature. Pathol Int 2012;62:817-22. [Crossref] [PubMed]
- Suda K, Tomizawa K, Yatabe Y, et al. Lung cancers unrelated to smoking: characterized by single oncogene addiction? Int J Clin Oncol 2011;16:294-305. [Crossref] [PubMed]
- Lee JJ, Park S, Park H, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177:1842-57.e21. [Crossref] [PubMed]
- Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402-7. [Crossref] [PubMed]
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77. [Crossref] [PubMed]
- Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27-40. [Crossref] [PubMed]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341-54. [Crossref] [PubMed]
- Huang HE, Chin SF, Ginestier C, et al. A recurrent chromosome breakpoint in breast cancer at the NRG1/neuregulin 1/heregulin gene. Cancer Res 2004;64:6840-4. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014;4:415-22. [Crossref] [PubMed]
- Dimou A, Camidge DR. Detection of NRG1 Fusions in Solid Tumors: Rare Gold? Clin Cancer Res 2019;25:4865-7. [Crossref] [PubMed]
- Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014;20:3087-93. [Crossref] [PubMed]
- Gow CH, Wu SG, Chang YL, et al. Multidriver mutation analysis in pulmonary mucinous adenocarcinoma in Taiwan: identification of a rare CD74-NRG1 translocation case. Med Oncol 2014;31:34. [Crossref] [PubMed]
- Dhanasekaran SM, Balbin OA, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun 2014;5:5893. [Crossref] [PubMed]
- Nagasaka M, Ou SI. Neuregulin 1 Fusion-Positive NSCLC. J Thorac Oncol 2019;14:1354-9. [Crossref] [PubMed]
- Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin Cancer Res 2019;25:4966-72. [Crossref] [PubMed]
- Ke H, Shen W, Hu A, et al. Distribution of NRG1 Gene Fusions in a Large Population of Chinese Patients with NSCLC. J Thorac Oncol 2019;14:e263-6. [Crossref] [PubMed]
- Pan Y, Zhang Y, Ye T, et al. Detection of Novel NRG1, EGFR, and MET Fusions in Lung Adenocarcinomas in the Chinese Population. J Thorac Oncol 2019;14:2003-8. [Crossref] [PubMed]
- Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol 2017;28:3092-7. [Crossref] [PubMed]
- Cheema PK, Doherty M, Tsao MS. A Case of Invasive Mucinous Pulmonary Adenocarcinoma with a CD74-NRG1 Fusion Protein Targeted with Afatinib. J Thorac Oncol 2017;12:e200-2. [Crossref] [PubMed]
- Gay ND, Wang Y, Beadling C, et al. Durable Response to Afatinib in Lung Adenocarcinoma Harboring NRG1 Gene Fusions. J Thorac Oncol 2017;12:e107-10. [Crossref] [PubMed]
- Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer Discov 2018;8:686-95. [Crossref] [PubMed]
- Schram A, O'Reilly E, Somwar R, et al. Clinical proof-of-concept for MCLA-128, a bispecific HER2/3 antibody therapy, in NRG1 fusion-positive cancers. Presented at the 2019 AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics in Boston, MA, October 26-30, 2019 2019.
- Kim HS, Han JY, Shin DH, et al. EGFR and HER3 signaling blockade in invasive mucinous lung adenocarcinoma harboring an NRG1 fusion. Lung Cancer 2018;124:71-5. [Crossref] [PubMed]
- Cho JH, Ku BM, Sun JM, et al. KIF5B-MET Gene Rearrangement with Robust Antitumor Activity in Response to Crizotinib in Lung Adenocarcinoma. J Thorac Oncol 2018;13:e29-31. [Crossref] [PubMed]
- Plenker D, Bertrand M, de Langen AJ, et al. Structural Alterations of MET Trigger Response to MET Kinase Inhibition in Lung Adenocarcinoma Patients. Clin Cancer Res 2018;24:1337-43. [Crossref] [PubMed]
- Qin A, Johnson A, Ross JS, et al. Detection of Known and Novel FGFR Fusions in Non-Small Cell Lung Cancer by Comprehensive Genomic Profiling. J Thorac Oncol 2019;14:54-62. [Crossref] [PubMed]
- Aggarwal C, Redman MW, Lara PN Jr, et al. SWOG S1400D (NCT02965378), a Phase II Study of the Fibroblast Growth Factor Receptor Inhibitor AZD4547 in Previously Treated Patients With Fibroblast Growth Factor Pathway-Activated Stage IV Squamous Cell Lung Cancer (Lung-MAP Substudy). J Thorac Oncol 2019;14:1847-52. [Crossref] [PubMed]
- Konduri K, Gallant JN, Chae YK, et al. EGFR Fusions as Novel Therapeutic Targets in Lung Cancer. Cancer Discov 2016;6:601-11. [Crossref] [PubMed]
- Zhu YC, Wang WX, Xu CW, et al. EGFR-RAD51 fusion variant in lung adenocarcinoma and response to erlotinib: A case report. Lung Cancer 2018;115:131-4. [Crossref] [PubMed]
- Raez LE, Pinto JA, Schrock AB, et al. EGFR-RAD51 Fusion: A Targetable Partnership Originated from the Tumor Evolution? J Thorac Oncol 2018;13:e33-4. [Crossref] [PubMed]
- Guan Y, Song Z, Li Y, et al. Effectiveness of EGFR-TKIs in a Patient with Lung Adenocarcinoma Harboring an EGFR-RAD51 Fusion. Oncologist 2019;24:1027-30. [Crossref] [PubMed]
- Zhu YC, Wang WX, Xu CW, et al. A Patient With Lung Adenocarcinoma With BRAF Gene Fusion and Response to Vemurafenib. Clin Lung Cancer 2019;20:e224-8. [Crossref] [PubMed]
- Shin DH, Jo JY, Han JY. Dual Targeting of ERBB2/ERBB3 for the Treatment of SLC3A2-NRG1-Mediated Lung Cancer. Mol Cancer Ther 2018;17:2024-33. [Crossref] [PubMed]
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- Gow CH, Liu YN, Li HY, et al. Oncogenic Function of a KIF5B-MET Fusion Variant in Non-Small Cell Lung Cancer. Neoplasia 2018;20:838-47. [Crossref] [PubMed]
- Song Z, Xu C, He Y, et al. Simultaneous Detection of Gene Fusions and Base Mutations in Cancer Tissue Biopsies by Sequencing Dual Nucleic Acid Templates in Unified Reaction. Clin Chem 2020;66:178-87. [Crossref] [PubMed]
- Zhu YC, Wang WX, Xu CW, et al. Identification of a novel crizotinib-sensitive MET-ATXN7L1 gene fusion variant in lung adenocarcinoma by next generation sequencing. Ann Oncol 2018;29:2392-3. [Crossref] [PubMed]
- Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231-5. [Crossref] [PubMed]
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795-803. [Crossref] [PubMed]
- Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636-47. [Crossref] [PubMed]
- Capelletti M, Dodge ME, Ercan D, et al. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res 2014;20:6551-8. [Crossref] [PubMed]
- Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res 2014;20:4107-14. [Crossref] [PubMed]
- Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [Crossref] [PubMed]
- Jang JS, Lee A, Li J, et al. Common Oncogene Mutations and Novel SND1-BRAF Transcript Fusion in Lung Adenocarcinoma from Never Smokers. Sci Rep 2015;5:9755. [Crossref] [PubMed]
- Lee NV, Lira ME, Pavlicek A, et al. A novel SND1-BRAF fusion confers resistance to c-Met inhibitor PF-04217903 in GTL16 cells through [corrected] MAPK activation. PLoS One 2012;7:e39653. [Crossref] [PubMed]
- Zhong R, Li H, Liu Y, et al. Chemotherapy combined with bevacizumab for the treatment of advanced lung adenocarcinoma cancer harboring EGFR-ANXA2, EGFR-RAD51, ATR and BRCA2 mutations: A case report. Thorac Cancer 2020;11:456-60. [Crossref] [PubMed]
- Jang YE, Jang I, Kim S, et al. ChimerDB 4.0: an updated and expanded database of fusion genes. Nucleic Acids Res 2020;48:D817-D824. [PubMed]
- Hibi M, Kaneda H, Tanizaki J, et al. FGFR gene alterations in lung squamous cell carcinoma are potential targets for the multikinase inhibitor nintedanib. Cancer Sci 2016;107:1667-76. [Crossref] [PubMed]
- Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol 2013;230:270-6. [Crossref] [PubMed]
- Bahleda R, Italiano A, Hierro C, et al. Multicenter Phase I Study of Erdafitinib (JNJ-42756493), Oral Pan-Fibroblast Growth Factor Receptor Inhibitor, in Patients with Advanced or Refractory Solid Tumors. Clin Cancer Res 2019;25:4888-97. [Crossref] [PubMed]
- Suda K, Mizuuchi H, Maehara Y, et al. Acquired resistance mechanisms to tyrosine kinase inhibitors in lung cancer with activating epidermal growth factor receptor mutation-diversity, ductility, and destiny. Cancer Metastasis Rev 2012;31:807-14. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Lin JJ, Ritterhouse LL, Ali SM, et al. ROS1 Fusions Rarely Overlap with Other Oncogenic Drivers in Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:872-7. [Crossref] [PubMed]
- Klempner SJ, Bazhenova LA, Braiteh FS, et al. Emergence of RET rearrangement co-existing with activated EGFR mutation in EGFR-mutated NSCLC patients who had progressed on first- or second-generation EGFR TKI. Lung Cancer 2015;89:357-9. [Crossref] [PubMed]
- Zhu VW, Klempner SJ, Ou SI. Receptor Tyrosine Kinase Fusions as an Actionable Resistance Mechanism to EGFR TKIs in EGFR-Mutant Non-Small-Cell Lung Cancer. Trends Cancer 2019;5:677-92. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Nelson AW, Schrock AB, Pavlick DC, et al. Novel SPECC1L-MET Fusion Detected in Circulating Tumor DNA in a Patient with Lung Adenocarcinoma following Treatment with Erlotinib and Osimertinib. J Thorac Oncol 2019;14:e27-9. [Crossref] [PubMed]
- Vojnic M, Kubota D, Kurzatkowski C, et al. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR therapy in EGFR-Mutated Lung Cancers. J Thorac Oncol 2019;14:802-15. [Crossref] [PubMed]
- Enrico D, Lacroix L, Chen J, et al. Oncogenic Fusions May Be Frequently Present at Resistance of EGFR Tyrosine Kinase Inhibitors in Patients With NSCLC: A Brief Report. JTO Clin Res Rep 2020;1:100023. [Crossref]
- Pros E, Saigi M, Alameda D, et al. Genome-wide profiling of non-smoking-related lung cancer cells reveals common RB1 rearrangements associated with histopathologic transformation in EGFR-mutant tumors. Ann Oncol 2020;31:274-82. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Boyle TA, Quinn GP, Schabath MB, et al. A community-based lung cancer rapid tissue donation protocol provides high-quality drug-resistant specimens for proteogenomic analyses. Cancer Med 2020;9:225-37. [Crossref] [PubMed]


