KRAS G12C mutations in Asia: a landscape analysis of 11,951 Chinese tumor samples
Introduction
Kirsten rat sarcoma vial oncogene (KRAS), member of the RAS superfamily, is one of the most prevalent oncogenes in cancer (1). Being a GTP-binding protein that links receptor tyrosine kinase activation to intracellular signalling, KRAS mutations favour the GTP-bound active state and constitutive activation of downstream effects including differentiation, proliferation and survival. Presence of KRAS mutations have been shown to be a negative prognostic factor in multiple cancer types including lung and colorectal cancers (2-5). In addition, presence of KRAS mutations is a predictive biomarker for EGFR-directed monoclonal antibodies in patients with colorectal cancer.
The most frequent mutations of KRAS occur at codon 12 (6), but the incidence of specific missense mutation at codon 12 are variable among different cancer types. For example, in non-small cell lung cancers (NSCLC), the most common KRAS mutation is G12C, whereas G12D is more common in pancreatic cancer (7,8). Incidence of KRAS mutations also differs between ethnic groups. Specifically, less than 10% of Asian patients with advanced NSCLC harbor KRAS mutation (9-11), while the incidence of KRAS mutations in African-Americans and Caucasians is 19% and 26%, respectively (12). Moreover, distribution of KRAS subtypes also varies between ethnic populations. Prior reports have shown KRAS G12C as the most common subtype amongst in African Americans (38%) and Caucasians (38%). A smaller cohort study of 218 KRAS Chinese NSCLC patients also reported G12C being the most common subtype, accounting for 32.1% of all KRAS mutations (9,12). Other reports have illustrated distinct subtypes of KRAS mutations between smokers and never smokers (13).
Recent report of a phase I study on AMG 510 is promising. This novel, first-in-class, small molecule specifically inhibits KRAS G12C by locking it in an inactive GDP-bound state (14,15). Tumor response rate in 23 patients with KRAS G12C positive NSCLC was 48% (16). Based on these preliminary results, the United States Food & Drug Administration has granted “fast-track” designation for AMG 510 (17). Moreover, other KRAS G12C specific inhibitors, including MRTX849 have also had promising initial early phase clinical trials data presented at international meetings (18). For future development of this class of agents, it is crucial to understand the comprehensive landscape of KRAS GI2C mutation across different tumor types, ethnicities and tobacco exposure. As RAS/RTK is a complex signalling pathway, co-existing genomic aberrations may impact on the clinical outcomes of KRAS G12C inhibition. In this study, we aim to study the epidemiologic landscape of KRAS G12C mutation in multiple cancer types in a large Chinese population and correlate the incidence of mutation with tobacco exposure in patients with NSCLC. Furthermore, we have also investigated the incidence of concomitant aberrations that may potentially impact on KRAS G12C inhibition. We present the following article in accordance with the STROBE reporting checklist (Available at http://dx.doi.org/10.21037/tlcr-20-455).
Methods
Patients and sample collection
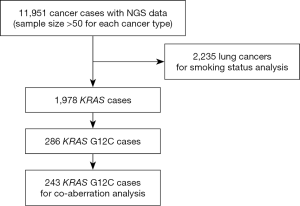
Total of 11,951 formalin-fixed, paraffin-embedded (FFPE) tumor tissue samples collected between 11/2016 and 7/2019 were analysed by next generation sequencing (NGS) (OrigiMed Ltd, Shanghai, China). This CAP-/CLIA-Accredited Laboratory offered three different types of gene panels commercially, all of which have all known coding exons of KRAS included in their analysis. For analysing concomitant aberrations, data from KRAS G12C samples were analysed, which were sequenced with Cancer Sequencing YS panel, a validated customized panel targeting over 450 cancer-related genes (19). All tumor samples were reviewed by in-house pathologists and only samples with 20% or more of tumor-cells cellularity were accepted for analysis. Smoking status and clinical characterization were available for 2,223 lung cancer cases within this cohort. Informed consent on plans of further deidentified genomic data analysis were obtained from all patients by test-ordering physicians as part of standard practice at respective institutions.
In order to obtain more representative data, only in diseases of which more than 100 samples have been received during the recruitment period are studied in this report.
All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). Due to its multi-institutional, anonymized and retrospective nature of data collection, in conjunction with subjects recruited in this study have already acknowledged and confirmed informed consent in proceeding with genomic testing and for the relevant anonymized used for further studies, ethical approval for this specific study has been waived.
Sequencing and detection of genomic alterations
For NGS, 50 to 200 ng DNA was extracted and purified from FFPE samples. Hybridization capture libraries were constructed and sequenced on Illumina sequencing platform (Illumina Incorporated, San Diego, CA), with a mean coverage of at least 700×. Genomic alterations were analysed with bioinformatics tools as reported previously (20). Single nucleotide variants (SNVs), short and long insertions/deletions (indels), copy number variations (CNVs) and gene fusions/rearrangements were analysed. For variant calling, at minimum five reads and variant allele frequency (VAF) of 1% were required.
COSMIC data
KRAS mutations were compared with Catalogue of Somatic Mutation In Cancer data (21) (COSMIC; https://cancer.sanger.ac.uk/cosmic, release v89, 15th May 2019), which provides access to publicly available genomic data of diverse cancers.
Identification of potentially actionable alterations by OncoKB
Four levels of evidence defined by MSK-Precision Oncology Knowledge base (OncoKB; https://oncokb.org/), were used to categorize potentially actionable alterations. Mutational events of each individual were annotated according to the OncoKB criteria.
Statistical analysis
R software was performed for statistical analyses. For comparing the frequency of KRAS mutation in Chinese population and COSMIC data, Chi-squared test was used to calculate the significance of differences in each cancer type separately. P value were adjusted through Benjamini and Hochberg (BH) procedure to control the false discovery rate (FDR). For analysing correlation between KRAS mutation and smoking status, Chi-squared test and fisher test was used to calculate the significance of differences. P value smaller than 0.05 were considered significant.
Results
Epidemiology of KRAS mutations
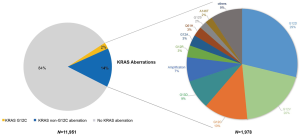
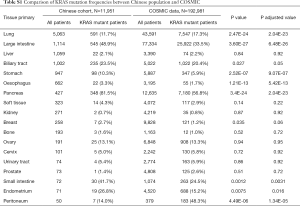
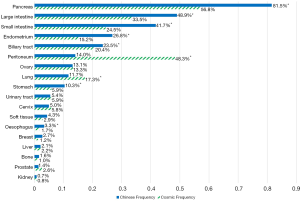
The most common cancer types in our study cohort included lung (42.4%), colorectal (9.3%), liver (8.9%), biliary tract (8.4%), stomach (7.9%), oesophagus (5.5%) and pancreas (3.6%; Table S1). We included only the cancer types with more than 50 cases for analysis, thus 11,951 cases were analysed (Figure 1). KRAS mutations were observed in 1,978 of 11,951 tumor samples (16.6%). Frequency of KRAS mutations varies between different cancer types, with highest frequencies observed in pancreatic (81.5%), colorectal (48.9%) and biliary tract (23.5%) cancer. Epidemiologic distribution was compared with the COSMIC database (22) (Figure 2). Incidence of KRAS mutation is higher in our Chinese patient cohort with pancreatic (81.5% vs. 56.8%; P<0.001), colorectal (48.9% vs. 33.5%; P<0.001) and gastric cancer (10.3% vs. 5.9%; P<0.001), while the incidence is lower in patients with lung cancer (11.7% vs. 17.3%; P<0.001).
Full table
KRAS G12C mutations
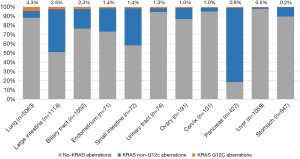
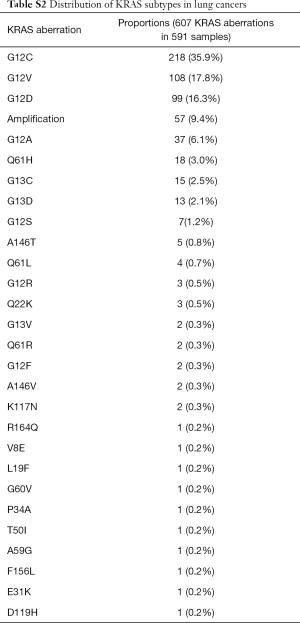
The majority of KRAS genomic aberrations were single nucleotide variations (SNVs), accounting for 91.9% of all KRAS alterations (Figure S1). Gene amplifications were the second most common type of alteration, accounting for a less proportion of only 7.1%. Among SNVs, G12C were detected in 286 samples, accounting for 14.5% of KRAS mutations and 2.4% of the entire study population. KRAS G12C mutation was more commonly found in lung, colorectal and biliary cancers (Figure 3). Out of 5,063 patients with lung cancer, 218 (4.3%) had KRAS G12C mutation and 373 (7.4%) had non-G12C KRAS mutations. Distribution of non-G12C KRAS mutations is summarized in Table S2. Similarly, 28 of 1,114 colorectal cancer patients (2.5%) had G12C mutation and 517 (46.4%) had non-G12C mutations; and 23 of 1,002 (2.3%) biliary cancer patients had G12C mutation and 212 (21.2%) had non-G12C mutations. Ratio of G12C versus non-G12C mutations was 1:1.7, 1:18.5 and 1:9 for lung, colorectal and biliary cancer, respectively. In contrast, only 4 of the 427 patients with pancreatic cancer had G12C mutation, the G12C versus non-G12C mutations ratio was 1:86.
Full table
Concomitant genomic aberrations in patients with KRAS G12C
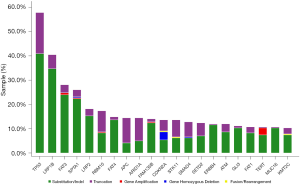
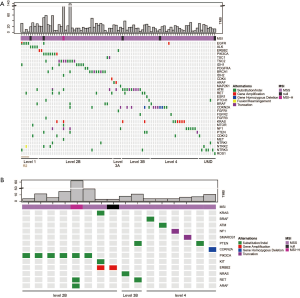
Total of 243 tumor samples with confirmed KRAS G12C were analysed with the comprehensive targeted gene panel. One or more concomitant aberrations were identified in 242 samples (99.5%). Median number of concomitant genomic aberrations was 14, ranging from 1 to 122. The most common concomitant aberration was TP53 (54.7%), LRP1B (37.0%) and FAT3 (25.1%; Figure 4). We have also identified 19 (7.8%) cases of co-existence of KRAS G12C and non-G12C mutations.
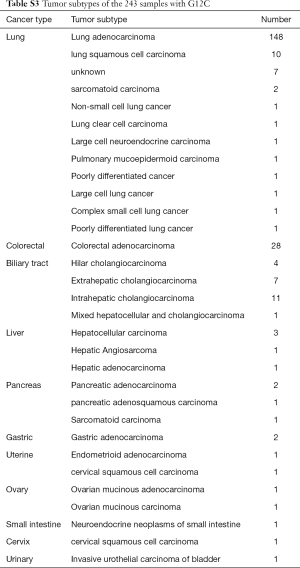
The histological subtypes of the 243 G12C tumors were further analysed (Table S3). The most common cancer histological subtype was lung adenocarcinoma (LUAD; N=148), followed by colorectal adenocarcinoma (CRC; N=28) and cholangiocarcinoma (CHOL; N=23). Frequencies of specific concomitant aberrations varied between different cancer subtypes. In LUAD, the most frequently altered genes were TP53 (50.0%), LRP1B (45.3%), and SPTA1 (30.4%; Figure S2A), comparing to CRC with most frequently altered genes at TP53 (71.4%), APC (53.6%), and FBXW7 (39.3%; Figure S2B). In CHOL, the most frequently co-altered genes were TP53 (60.9%), SMAD4 (39.1%), and CDKN2A (34.8%; Figure S2C). Given the number of G12C was low in other cancer histological subtypes, we did not analyse the concomitant aberration of this group.
Full table
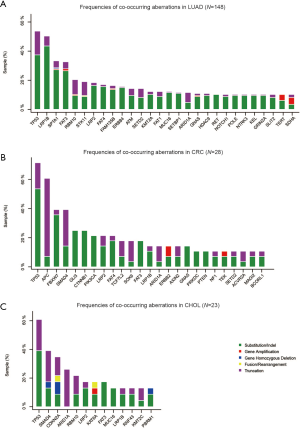
A Recent large-cohort-study reported detailed driver genes in different cancer types (23). The list of driver genes was obtained as reported in Table S4. Concomitant aberrations on driver genes in G12C positive LUAD, CRC and CHOL were analysed respectively (Figure 5). The most common co-occurring driver gene in LUAD was TP53 (50.6%), followed by RBM10 (19.6%) and STK11 (18.2%). EGFR aberrations occurred in 8.8% of cases. We also observed co-occurring KRAS non-G12C aberrations in 10 cases (6.8%), in which eight were KRAS amplification. In CRC, the most common co-occurring driver gene was TP53 (71.4%), followed by APC (53.6%) and FBXW7 (39.3%). Coaberrations in three driver genes were found in CHOL, which were ARID1A (26.1%), PBRM1 (13.0%), and EPHA2 (4.4%).
Full table

Smoking history and KRAS mutations
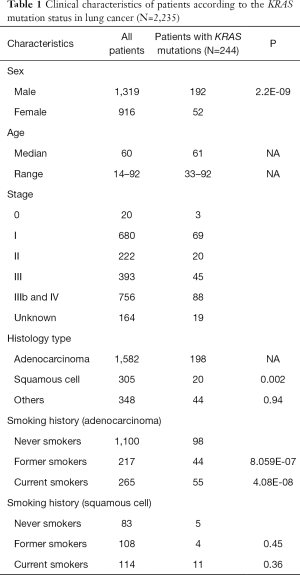
Smoking history of 2,235 lung cancer cases was collected. This included 1,582 LUAD and 305 squamous cell carcinoma histologies (Table 1). In LUAD, KRAS mutations were identified in 20.3% of former smokers, which were significantly higher than that of non-smokers (8.9%; P<0.001; Table 1). And in 20.7% of current smokers, significantly higher than non-smokers (P<0.001). On the contrary, smoking history did not play a significant role in the incidence of KRAS mutations in patients with lung squamous cell carcinomas (LUSC). Further details on KRAS mutation subtypes and respective correlation with smoking status were illustrated in Table S5.
Full table

Full table
Pathway analysis of co-occurring aberrations
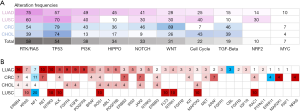
Prior studies indicated there were multiple canonical oncogenic pathways among diverse tumor types (24). Concomitant genes associated oncogenic pathways were analysed accordingly (Figure 6A). Frequency of mutations in each pathway differed according to tumor subtypes. For patients with LUAD, the most common and impactful oncogenic pathway was RTK/RAS signalling pathway, occurring in 75% of the G12C patients. For patients with CRC, the most impactful oncogenic pathway was WNT signalling pathway (89%). While for patients with CHOL and LUSC, the most common and impactful pathway was TP53-associated genes (74% in CHOL and 70% in LUSC). We explored the profile of genomic mutations of RTK/RAS pathway-associated genes (Figure 6B) and identified 35 key genes in associated with KRAS mutation, and among which, three were tumor suppressor genes.
Most of these identified genes function upstream of the KRAS pathway, while several downstream signalling molecules were also observed (Figure 7A). Concomitant non-G12C KRAS mutations were observed in LUAD and CRC, occurred in 6.8% and 3.8% respectively. Preclinical data showed that PI3K-AKT pathway inactivation was likely intrinsic resistance mechanism for G12C inhibitors (25). BRAF mutations would provide fitness advantage for subclones resistant to G12C inhibition (26). Associated genes on the pathway were analysed in LUAD and CRC. BRAF is on the downstream of KRAS in RTK/RAS pathway. The incidence of BRAF mutation in G12C mutated patients were comparable between LUAD and CRC, occurring in 5.4% and 3.6% patients respectively (P=0.9; Figure 7A). PI3K pathway is downstream signalling of RAS. In this study, a higher proportion of PI3KCA mutation was observed in CRC (21.4% vs. 8.8%; P=0.04; Figure 7B).
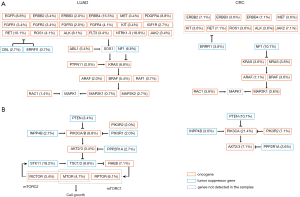
Actionable co-alterations analysis
Potential actionable co-alterations were analysed as defined by the OncoKB classification (27). Level 1 actionable alteration was found only in LUAD (Table 2). In G12C LUAD cases (Figure S3A), 81 (54.7%) cases had at least one potentially actionable alteration in addition to KRAS G12C (level 1 to 4). Actionable EGFR and ALK mutations were identified in eight cases (5.4%), which were recognized as FDA-approved biomarkers for target therapies (level 1). Other cases can also be potentially targetable either with off-label drugs or therapies that are currently in clinical trials (level 2B to 4). In colorectal carcinomas (Figure S3B), 16 were potentially targetable (57.1%; level 2B to 4). In total, 52.3% of G12C patients with co-aberrations had at least one actionable alteration.
Full table
Discussion
Recent discoveries have provided promising therapeutic opportunities for patients harboring KRAS G12C mutations. To the best of our knowledge, our report is the largest single-cohort illustrating the genomic and epidemiological landscape of KRAS G12C mutations in cancer. Moreover, this dataset represents the largest cohort of Chinese cancer patients with tumors harboring KRAS mutations ever assembled.
Higher frequencies of KRAS mutation were observed in several common cancers, including colorectal, stomach and pancreatic cancer in our study than reported in COSMIC, most of which were data from western population. This discrepancy may possibly be due to the higher resolution technology being used in our study which mean that mutations with low variation frequency (VAF) were also detectable. In the current study, the full exon of KRAS was detected using NGS with a mean coverage of at least 700×, at minimal VAF of 1% could be detected in this study.
On the contrary, the frequency of KRAS mutations were lower in our Chinese lung cancer cohort when compared with Western series. Similar findings have been reported in prior studies of Chinese populations (9,20). The imbalance of prevalence of molecular drivers in LUAD between Asian and Caucasian populations has been well documented. Given the fact that EGFR mutations are more common in Asians, it is not inconceivable that the relative prevalence of other molecular drivers, including KRAS mutations, are altered. The lower proportion of KRAS mutations in Asian LUADs in general may explain the overall lower frequency of KRAS G12C compared with previous reports among Western populations in lung cancer (7,28). In colorectal and pancreatic cancer patients, the frequencies of KRAS G12C were comparable with prior reports (29,30). Interestingly, 2.3% of biliary tract cancer patients were observed with KRAS G12C in our dataset. This information on the molecular landscape of KRAS G12C mutations may have significant impact on operational aspects in conducting clinical trials with KRAS G12C specific inhibitors in the Chinese population.
Moreover, presence of possible co-occurrence of other aberrations aside from KRAS G12C were evaluated in all patients within this study. The frequencies of co-altered genes were different in cancers. This has provided us with valuable insight in identifying potential pathways of treatment resistance in patients who are to be treated with KRAS G12C specific inhibitors, as the mechanism of resistance to these inhibitors may have already been established de-novo.
The presence of KRAS mutation suggests lack of response to EGFR targeted therapies in non-small lung cancers and colorectal cancers (31,32). As KRAS downstream of EGFR in the RAS/RTK pathway, there may be potential to consider combination therapies in the rare patients who harbor both of these alterations, or develop KRAS mutations in tumors as secondary resistance to EGFR inhibitors. 52.3% of patients with co-aberrations had at least one actionable alteration in accordance to the OncoKB definition, only 3.3% of these patients had level 1 co-aberrations which were targetable by FDA-approved therapies.
Preliminary data on AMG 510 showed clinical efficacy on KRAS G12C mutated NSCLC but not CRC. This might be intrinsic mechanism underlying the difference. Although the genomic mechanisms of intrinsic resistance to kinase inhibitors are complicated, there are usually two categories. One is the secondary mutation on the targeting kinase, second is activation of other molecules on the downstream of the pathway. In the current study, aberrant RAS/RTK pathway were broadly observed in KRAS G12C tumors. Downstream genomic alterations, such as RAFs and PI3Ks alterations, may increase the risk of drug resistance to KRAS G12C inhibitors. This is in line with a recent study which demonstrated the presence of BRAF mutations leading to primary resistance to G12C inhibitors (26). Preclinical study is also supportive of the observation of PI3K-AKT as reason of intrinsic resistance to G12C inhibitors (25). The fact that we observed high incidence of PI3KCA mutation in patients with CRC may potentially explain the lack of response in this patient group. Considering the genomic difference between populations in study, further investigations are still needed.
This is a retrospective analysis of Real-World data based on a commercial platform; thus, we have limited clinical information from original source documents. Information pertaining to patients’ demographics and clinical information were based on the test request form. We are limited by the lack survival outcomes for correlation with the mutation status. However, the large sample collection on a relatively homogenous ethnic population by an identical platform has provided us with valuable and important data for further investigations. It will be extremely challenging to conduct a perspective study with similar sample size. But considering the relative rarity of KRAS G12C mutation, a large sample size is mandatory for accurate evaluation.
Our study has demonstrated the genomic landscape of KRAS G12C mutations in a large Chinese population and we confirmed the incidence to be relatively low. The most common tumor types harboring the mutation are lung, colorectal and biliary cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-455
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-455). HHFL reports personal fees from Novartis, personal fees from Pfizer, grants and personal fees from MSD, personal fees from Eisai, personal fees from Boehringer-Ingelheim, grants from Mundipharma, outside the submitted work; CC, HL, JY, YX, MY, KW reports employment by Origimed, during the conduct of the study; TSKM reports grants and personal fees from AstraZeneca, grants and personal fees from Roche/Genentech, personal fees from Eli Lilly, grants and personal fees from BMS, grants and personal fees from Boehringer Ingelheim, grants and personal fees from Novartis, grants and personal fees from MSD, grants and personal fees from Pfizer, personal fees from Merck Serono, grants and personal fees from Clovis Oncology, personal fees from Vertex, grants and personal fees from SFJ Pharmaceuticals, personal fees from ACEA Biosciences, from geneDecode, personal fees from Oncogenex, personal fees from Celgene, personal fees from Ignyta Inc, grants and personal fees from Taiho, grants from Eisai, personal fees from Fishawack Facilitate Ltd, grants and personal fees from Takeda, personal fees from Janssen, personal fees from Hutchison ChiMed, grants from XCovery, personal fees from OrigiMed, personal fees from Hengrui Therapeutics, personal fees from Sanofi-Aventis R&D, personal fees from Yuhan Corporation, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). Due to its multi-institutional, anonymized and retrospective nature of data collection, in conjunction with subjects recruited in this study have already acknowledged and confirmed informed consent in proceeding with genomic testing and for the relevant anonymized used for further studies, ethical approval for this specific study has been waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haigis KM. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017;3:686-97. [Crossref] [PubMed]
- Martin P, Leighl NB, Tsao MS, et al. KRAS mutations as prognostic and predictive markers in non-small cell lung cancer. J Thorac Oncol 2013;8:530-42. [Crossref] [PubMed]
- Blons H, Emile JF, Le Malicot K, et al. Prognostic value of KRAS mutations in stage III colon cancer: post hoc analysis of the PETACC8 phase III trial dataset. Ann Oncol 2014;25:2378-85. [Crossref] [PubMed]
- Hecht JR, Cohn A, Dakhil S, et al. SPIRITT: A Randomized, Multicenter, Phase II Study of Panitumumab with FOLFIRI and Bevacizumab with FOLFIRI as Second-Line Treatment in Patients with Unresectable Wild Type KRAS Metastatic Colorectal Cancer. Clin Colorectal Cancer 2015;14:72-80. [Crossref] [PubMed]
- Tosi F, Magni E, Amatu A, et al. Effect of KRAS and BRAF Mutations on Survival of Metastatic Colorectal Cancer After Liver Resection: A Systematic Review and Meta-Analysis. Clin Colorectal Cancer 2017;16:e153-63. [Crossref] [PubMed]
- Stolze B, Reinhart S, Bulllinger L, et al. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep 2015;5:8535. [Crossref] [PubMed]
- Scheffler M, Ihle MA, Hein R, et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J Thorac Oncol 2019;14:606-16. [Crossref] [PubMed]
- Pantsar T, Rissanen S, Dauch D, et al. Assessment of mutation probabilities of KRAS G12 missense mutants and their long-timescale dynamics by atomistic molecular simulations and Markov state modeling. PLoS Comput Biol 2018;14:e1006458. [Crossref] [PubMed]
- Jia Y, Jiang T, Li X, et al. Characterization of distinct types of KRAS mutation and its impact on first-line platinum-based chemotherapy in Chinese patients with advanced non-small cell lung cancer. Oncol Lett 2017;14:6525-32. [Crossref] [PubMed]
- Gao B, Sun Y, Zhang J, et al. Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J Thorac Oncol 2010;5:1130-5. [Crossref] [PubMed]
- Xia N, An J, Jiang QQ, et al. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res 2013;39:328-35. [Crossref] [PubMed]
- Reinersman JM, Johnson ML, Riely GJ, et al. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in African Americans. J Thorac Oncol 2011;6:28-31. [Crossref] [PubMed]
- Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169-77. [Crossref] [PubMed]
- AMG 510 First to Inhibit "Undruggable" KRAS. Cancer Discov 2019;9:988-9. [PubMed]
- Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217-23. [Crossref] [PubMed]
- Govindan R, Fakih M, Price T, et al. editors. Phase 1 study of safety, tolerability, PK and efficacy of AMG 510, a novel KRASG12c inhibitor, evaluated in NSCLC. World Conference on Lung Cancer 2019; 2019 September 8, 2019; Barcelona, Spain.
- Amgen Announces New Clinical Data Evaluating Novel Investigational KRAS(G12C) Inhibitor In Larger Patient Group At WCLC 2019. Available online: https://www.amgen.com/media/news-releases/2019/09/amgen-announces-new-clinical-data-evaluating-novel-investigational-krasg12c-inhibitor-in-larger-patient-group-at-wclc-2019/
- Janne PA, Papadopoulos K, Ou I, et al. A phase 1 clinical trial evaluating the pharmacokinetics (PK), safety, and clinical activity of MRTX849, a mutant-selective small molecule KRAS G12C inhibitor, in advanced solid tumors. AACR-NCI-EORTC International Conference on Molecular Targets; October 28, 2019.
- Cao J, Chen L, Li H, et al. An Accurate and Comprehensive Clinical Sequencing Assay for Cancer Targeted and Immunotherapies. Oncologist 2019;24:e1294-302. [Crossref] [PubMed]
- Wen S, Dai L, Wang L, et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncologist 2019;24:e1070-81. [Crossref] [PubMed]
- Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 2004;91:355-8. [Crossref] [PubMed]
- Margonis GA, Kim Y, Spolverato G, et al. Association Between Specific Mutations in KRAS Codon 12 and Colorectal Liver Metastasis. JAMA Surg 2015;150:722-9. [Crossref] [PubMed]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;174:1034-5. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-37.e10. [Crossref] [PubMed]
- Misale S, Fatherree JP, Cortez E, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 2019;25:796-807. [Crossref] [PubMed]
- Cannataro VL, Gaffney SG, Stender C, et al. Heterogeneity and mutation in KRAS and associated oncogenes: evaluating the potential for the evolution of resistance to targeting of KRAS G12C. Oncogene 2018;37:2444-55. [Crossref] [PubMed]
- Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017. [Crossref] [PubMed]
- Arbour KC, Jordan E, Kim HR, et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:334-40. [Crossref] [PubMed]
- Zhou L, Baba Y, Kitano Y, et al. KRAS, BRAF, and PIK3CA mutations, and patient prognosis in 126 pancreatic cancers: pyrosequencing technology and literature review. Med Oncol 2016;33:32. [Crossref] [PubMed]
- Neumann J, Zeindl-Eberhart E, Kirchner T, et al. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract 2009;205:858-62. [Crossref] [PubMed]
- Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008;9:962-72. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]