
Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: a pilot study
Introduction
The risk of post-surgical recurrence is still problematic even when locoregional control is thought to have been achieved by complete surgical resection. For example, a Japanese lung cancer registry study (n=18,973) reported that the disease-free survival rate at 5 years after pulmonary resection was 67.8% (1). To improve the outcomes of surgically resected non-small cell lung cancer (NSCLC) patients, several clinical trials employing tyrosine kinase inhibitors (2) or new chemotherapeutic regimen (3) have been performed. In addition to these efforts to develop novel adjuvant therapies, evaluation of the personal risks of recurrence is an important issue for a better post-surgical care (4), including the detection of post-surgical minimal residual disease (MRD) (5). If we are able to exclude patients who do not relapse from the candidates of adjuvant therapy, it will eliminate unnecessary adverse events or costs.
Circulating tumor DNA (ctDNA) analysis, which detects mutations of circulating cell-free DNA (cfDNA) that is shed by tumor cells, is a useful technique for detecting mutations in tumor cells. Detecting mutations in EGFR, including the T790M resistant mutation, is already used for clinical decision-making in advanced NSCLC patients (6). It has been reported that ctDNA analysis prior to the surgery in early-stage lung cancer patients can detect somatic mutations in tumors at high sensitivity and specificity (7), and that ctDNA analysis is able to detect mutations which will present even in a heterogeneous manner in tumor tissues (8,9). Additionally, recent studies have suggested that ctDNA can be a potential biomarker for the assessment of post-surgical MRD (10), as well as a potential predictor for the disease progression prior to radiological modalities (11).
In early-stage lung cancer patients, tumor markers are only available by blood tests and are used to estimate tumor burden after pulmonary resection. Although some retrospective analyses have reported that some tumor markers are prognostic factors (12,13), none are currently recommended as a biomarker to select patients with poorer prognosis in any lung cancer treatment guidelines. Therefore, several groups have evaluated the clinical implication of more sensitive indicators, among which ctDNA detection is a useful candidate (11,14-22). A recent study also reported that ctDNA levels rapidly decreased after radical tumor resection, and that ctDNA levels between postoperative day 3 (POD 3) through 1 month (POD 1M) could be used as baseline values for postoperative lung cancer surveillance (20).
Despite the many reports of ctDNA detection in surgically resected lung cancer patients, it is unclear if ctDNA detection is superior to testing tumor markers. Additionally, it is unclear if ctDNA analysis should be performed postoperatively, preoperatively, or both. To answer these questions, we performed this prospective pilot study on 20 clinical stage IIA–IIIA lung cancer patients. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-505).
Methods
Study cohort
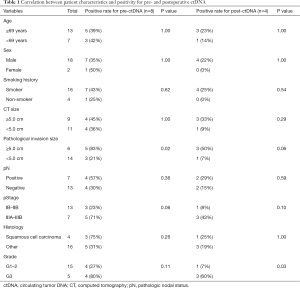
Between January 2018 and May 2019, 23 lung cancer patients with clinical stage IIA–IIIA disease who underwent complete surgical resection (inclusion criteria) were intended to be included in this study. Patients who received neoadjuvant chemotherapy or had advanced malignancies other than lung cancer within the past 5 years were excluded (exclusion criteria). Among these 23 patients, one patient without enough plasma sample and two patients who refused the enrollment were excluded from the study. Finally, the data of 20 patients were analyzed in this study. The cohort consisted of 18 men and 2 women with a median age of 69 (range, 37–88) years; there were four squamous cell carcinomas, 13 adenocarcinomas, two adenosquamous cell carcinomas, and one small cell carcinoma. The details of patient backgrounds are summarized in Table 1. Seven patients received adjuvant chemotherapy and 13 did not. This study was approved by the Institutional Review Board of Kindai University Faculty of Medicine [30-009]. Written informed consent was obtained from each patient. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Full table
Mutation analysis of primary tumor samples
For the primary tumor tissues, mutations in the 409 cancer-related genes were analyzed using an Ion AmpliSeq Library Kit 2.0 and Ion AmpliSeq™ Comprehensive Cancer Panel (CCP) (Thermo Fisher Scientific, Waltham, MA, USA). Tumor DNA was extracted using All Prep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) and 40 ng of DNA were subjected to multiplex PCR amplification. Then, the Ion Xpress Barcode Adapters (Thermo Fisher Scientific) were ligated to the PCR products, which were then purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). The purified libraries were pooled, and then sequenced with the Ion Torrent S5 instrument and Ion 550 Chip Kit (both from Thermo Fisher Scientific). DNA sequencing data were accessed through the Torrent Suite v.5.10 program (Thermo Fisher Scientific). Reads were aligned against the hg19 human reference genome, and variants were called using Variant Call Format ver. 5.10. Raw variant calls were filtered by quality score <100, depth coverage <19, and were manually checked using the integrative genomics viewer (IGV; Broad Institute, Cambridge, MA, USA). Germline mutations were excluded using the Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB) and the Exome Aggregation Consortium database.
Extraction and detection of cell-free DNA (cfDNA)
Blood samples were collected in cell-free ctDNA tubes (Roche Diagnostics, Mannheim, Germany) and stored at room temperature. Samples were processed within 1 week of collection by centrifuging the blood for 10 min at 1,600 ×g. The plasma was stored at −80 °C until use. The entire plasma sample [up to 4 mL per case (range, 3–4 mL)] was used for cfDNA extraction. Plasma cfDNA was purified using an AVENIO cfDNA isolation kit (Roche Diagnostics), according to the manufacturer’s instructions. The quality and quantity of the DNA were verified using a NanoDrop 2000 device (Thermo Fisher Scientific) and the PicoGreen dsDNA assay kit (Life Technologies, Carlsbad, CA, USA). Extracted cfDNA samples were stored at −80 °C until analysis.
In eight patients, preoperative blood sampling was performed within 24 h before surgery and in 12 patients, preoperative blood sampling was performed between 24–48 h before surgery. Postoperative blood sampling was performed within an average of 6.3 (range, 3–12) days after surgery. We detected ctDNA in plasma using the cancer personalized profiling by deep sequencing (CAPP-Seq) technique, which can detect mutations in 197 genes (Roche Diagnostics). We counted mutations as positive when a detected single nucleotide variant had been previously reported in lung cancers in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (https://cancer.sanger.ac.uk/cosmic).
Statistical analysis
Fisher’s exact test was used to compare differences in categorical variables. Recurrence-free survival (RFS) was defined as the interval between the date of surgery and the date of recurrence or death by any cause. RFS was estimated by the Kaplan-Meier method and compared using the log rank test. A P value <0.05 was considered statistically significant. All data were analyzed using JMP version 13.0 software (SAS Institute, Cary, NC, USA).
Results
Comparison of mutational status between primary tumors and cfDNA
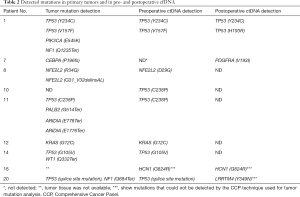
Mutation(s) in cfDNA were detected in nine patients from pre- and/or postoperative plasma samples (Table 2). Among the eight patients who had mutation data for the primary tumors, we observed that many of mutations detected in the primary tumors were also detectable in cfDNA (all five TP53 mutations and one KRAS G12C mutation). However, there were also discrepancies between the mutational status of primary tumors and cfDNA samples, for example, two TP53 mutations (H193R in patient 2 and C238P in patient 15) that were detected in preoperative ctDNA were not identified in primary tumors. Additionally, a PDGFRA (V193I) mutation that was detected in postoperative ctDNA was not identified in the primary tumor or preoperative ctDNA. These discrepancies may indicate the presence of intratumor genetic heterogeneity.
Full table
Correlation between clinicopathological factors and pre-/postoperative ctDNA detection
The patient characteristics are summarized in Table 1. Eight patients (40%) were positive for preoperative ctDNA, and preoperative ctDNA positivity was significantly correlated with pathological tumor size (≥5.0 cm, P=0.018). Four patients (20%) were positive for postoperative ctDNA, and postoperative ctDNA positivity was significantly correlated with histological Grade 3 (P=0.032), while pathological tumor size did not reach statistical significance (P=0.061).
Prognostic value of ctDNA analysis
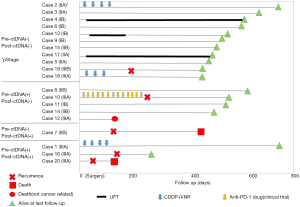
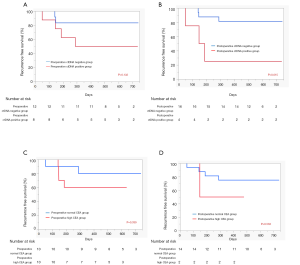
Next, we evaluated the prognostic implications of pre- and postoperative ctDNA positivity using RFS as the endpoint. At the time of data analysis, the 20 patients had been monitored for a median follow-up of 12 months. Detailed ctDNA status, pathological stage, adjuvant therapy, and survival data of each patient are shown in Figure 1. Among the 20 patients, five experienced recurrence and two died from lung cancer. In the RFS analysis, patients positive for preoperative ctDNA tended to have worse prognosis (P=0.132, Figure 2A). However, patients positive for postoperative ctDNA had significantly poorer prognoses compared with patients who were negative for postoperative ctDNA (P=0.015, Figure 2B). As expected, RFS was also significantly correlated with pathological stage (P=0.002, data not shown). However, histologic grade, which was significantly correlated with postoperative ctDNA positivity, was not a significant prognostic factor (P=0.49, data not shown). We also evaluated the prognostic value of pre- and postoperative CEA levels. Among the 20 patients in this study, all had preoperative CEA data and 16 had postoperative CEA data at 2–4 months after surgery. We found that neither preoperative (Figure 2C) nor postoperative (Figure 2D) CEA levels were not prognostic factors in our cohort (P=0.339 and P=0.533, respectively). Due to the small number of patients positive for postoperative ctDNA (n=4), we did not perform multivariate analysis.
Discussion
There are several techniques to detect ctDNA, including PCR-based (7,23), NGS panel based, and be spoked NGS (10). In this study, using a next-generation sequencing (NGS)-based panel test for cfDNA analysis, we found that detecting mutations in cancer-related genes from postoperative ctDNA (POD 3–12) predicted poor patient outcomes. This finding was compatible with a recent study by Chen et al. (20) that showed postoperative ctDNA detection (POD 3 or POD 1 month) was significantly correlated with RFS. Although the study by Chen et al. detected mutations in only nine cancer-related genes (EGFR, KRAS, ERBB2, BRAF, PIK3CA, ALK, RET, MET exon 14 skipping, and TP53), using an NGS-based detection platform, our study revealed that detecting other lung cancer-related genes (such as PDGFRA, HCN1, and LRRTM4) from ctDNA can also be prognostic. Therefore, we consider that postoperative cfDNA analysis with a comprehensive genetic panel will help to identify patients who may have high risk of post-surgical recurrence.
It is noteworthy that three of the four patients who were positive for postoperative ctDNA relapsed within 6 months. This result was similar to the observation by Hu et al. that two patients with detectable EGFR mutations in ctDNA at 1 month after surgery both experienced recurrence within 4 months of surgery (19). Although this study was a pilot study with a small cohort, our results also suggested that postoperative ctDNA detection may more accurately predict poor patient outcomes than the pre- or postoperative tumor marker analysis and preoperative ctDNA detection.
Conversely, another recent study by Isaksson et al. (17) reported that preoperative ctDNA was a predictor of recurrence in a cohort of 58 patients. Because positive preoperative ctDNA is correlated with tumor size, as shown in our result and by others (20-22), and with lymph node involvement, as shown by others (20,21), we consider preoperative ctDNA positivity to be correlated with advanced disease, but not a simple predictor of recurrence. In fact, the study by Isaksson et al. included 34 stage I patients, which were excluded in our cohort.
In conclusion, this study suggested that postoperative ctDNA positivity is a surrogate for MRD, meaning positivity for ctDNA postoperatively may predict early recurrence in patients with resected NSCLC. We anticipate that postoperative ctDNA analysis will be useful for formulating postoperative follow-up plans and determining the indications for postoperative adjuvant therapy; however, additional prospective studies with larger cohorts are needed to validate the practicability and economic efficacy of using liquid biopsy to predict recurrence.
Acknowledgments
We thank James P. Mahaffey, PhD, from Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript.
Funding: This study was supported by a research grant from the Uehara Memorial Foundation (Suda K).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-505
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-505
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-505
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-505). TM serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. Dr. Suda reports personal fees from AstraZeneca, grants and personal fees from Boehrinnger-Ingelheim, grants from Rain Therapeutics, outside the submitted work. Dr. Sakai reports personal fees from Roche Diagnostics, Bio-Rad, SRL Diagnostics, AstraZeneca, and Chugai Pharmaceutical, outside the submitted work. TF reports personal fees from Novartis, grants from Apollomics.Inc., outside the submitted work. KN reports grants and personal fees from Otsuka Pharmaceutical, Life Technologies Japan, Boehringer Ingelheim, and Eli Lilly; personal fees from Chugai Pharmaceutical, Eisai, from Pfizer, Novartis, MSD, Ono Pharmaceutical, Bristol-Myers Squibb, SymBio Pharmaceuticals Limited, Solasia Pharma, Yakult Honsha, Roche Diagnostics, AstraZeneca, Sanofi, Guardant Health, Takeda, and Kobayashi Pharmaceutical, grants from Ignyta, and Astellas, outside the submitted work. TM reports grants and personal fees from AstraZeneca, Boehrinnger-Ingelheim, and Pfizer, personal fees from MSD, grants and personal fees from Chugai, grants and personal fees from Ono, personal fees from Eli-Lilly, grants and personal fees from Daiiichi-Sankyo, personal fees from ThermoFisher, personal fees from Guardant, grants and personal fees from Taiho, personal fees from Amgen, and Novartis, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Institutional Review Board of Kindai University Faculty of Medicine [30-009]. Written informed consent was obtained from each patient. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Okami J, Shintani Y, Okumura M, et al. Demographics, Safety and Quality, and Prognostic Information in Both the Seventh and Eighth Editions of the TNM Classification in 18,973 Surgical Cases of the Japanese Joint Committee of Lung Cancer Registry Database in 2010. J Thorac Oncol 2019;14:212-22.
- Herbst RS, Tsuboi M, John T, et al. Osimertinib as adjuvant therapy in patients (pts) with stage IB–IIIA EGFR mutation positive (EGFRm) NSCLC after complete tumor resection: ADAURA. J Clin Oncol 2020;38:(suppl; abstr LBA5).
- Kenmotsu H, Yamamoto N, Yamanaka T, et al. Randomized Phase III Study of Pemetrexed Plus Cisplatin Versus Vinorelbine Plus Cisplatin for Completely Resected Stage II to IIIA Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol 2020;38:2187-96. [Crossref] [PubMed]
- Suda K. Personalized post-surgical care?-possible strategies for NSCLCs with EGFR mutation. Transl Lung Cancer Res 2020;9:441-5. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Zugazagoitia J, Gómez-Rueda A, Jantus-Lewintre E, et al. Clinical utility of plasma-based digital next-generation sequencing in oncogene-driven non-small-cell lung cancer patients with tyrosine kinase inhibitor resistance. Lung Cancer 2019;134:72-8. [Crossref] [PubMed]
- Leung M, Freidin MB, Freydina DV, et al. Blood-based circulating tumor DNA mutations as a diagnostic and prognostic biomarker for lung cancer. Cancer 2020;126:1804-9. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577-86. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Yamazaki M, Ishikawa H, Kunii R, et al. A combination of preoperative CT findings and postoperative serum CEA levels improves recurrence prediction for stage I lung adenocarcinoma. Eur J Radiol 2015;84:178-84. [Crossref] [PubMed]
- Nonaka M, Kataoka D, Yamamoto S, et al. Pre- and post-operative serum carcinoembryonic antigen in primary lung adenocarcinoma. Ann Thorac Cardiovasc Surg 2004;10:281-4. [PubMed]
- Jamal-Hanjani M, Hackshaw A, Ngai Y, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol 2014;12:e1001906. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [Crossref] [PubMed]
- Isaksson S, George AM, Jönsson M, et al. Pre-operative plasma cell-free circulating tumor DNA and serum protein tumor markers as predictors of lung adenocarcinoma recurrence. Acta Oncol 2019;58:1079-86. [Crossref] [PubMed]
- Liang H, Huang J, Wang B, et al. The role of liquid biopsy in predicting post-operative recurrence of non-small cell lung cancer. J Thorac Dis 2018;10:S838-45. [Crossref] [PubMed]
- Hu W, Yang Y, Zhang L, et al. Post surgery circulating free tumor DNA is a predictive biomarker for relapse of lung cancer. Cancer Med 2017;6:962-74. [Crossref] [PubMed]
- Chen K, Zhao H, Shi Y, et al. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin Cancer Res 2019;25:7058-67. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Ohira T, Sakai K, Matsubayashi J, et al. Tumor volume determines the feasibility of cell-free DNA sequencing for mutation detection in non-small cell lung cancer. Cancer Sci 2016;107:1660-6. [Crossref] [PubMed]
- Alama A, Coco S, Genova C, et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated with Nivolumab. J Clin Med 2019;8:1011. [Crossref] [PubMed]


