
Low T790M relative allele frequency indicates concurrent resistance mechanisms and poor responsiveness to osimertinib
Introduction
The treatment of patients with non-small cell lung cancer (NSCLC) has been revolutionized by the development of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) (1-5). For patients harboring EGFR-sensitizing mutations treated with first-generation (gefitinib/erlotinib) or second-generation EGFR-TKIs (afatinib/dacomitinib), the objective response rates (ORRs) range from 56% to 74%, which are significantly higher than those of patients receiving first-line platinum-based doublet chemotherapy (1,2,5). However, drug resistance eventually develops, which requires subsequent treatment with third-generation EGFR TKIs or chemotherapy. Approximately 50% of patients with an EGFR-sensitizing mutation develop a secondary mutation in EGFR at position 790 (T790M), which impairs the binding of TKIs to the EGFR kinase domain, leading to EGFR-TKI resistance (6). EGFR-TKI resistance may result in a median progression-free survival (PFS) of less than 1 year (3,4). Osimertinib, a 3rd generation irreversible EGFR-TKI designed to overcome such resistance, has been approved by the US Food and Drug Administration for the treatment of metastatic EGFRT790M+ NSCLC patients who have experienced disease progression from prior EGFR-TKI therapy (7). EGFR T790M testing has become a standard procedure when patients develop resistance to first- or second-generation EGFR-TKIs (8).
Due to patients’ unwillingness to undergo re-biopsy, or risk of biopsy complications (included pneumothorax and pulmonary hemorrhage), obtaining tissue samples remains challenging (9). Thus, minimally invasive liquid biopsy, which captures circulating biomarkers in patient fluid samples such as plasma, urine, and saliva using Cobas, digital PCR (ddPCR), or next-generation sequencing (NGS), has rapidly developed as an alternative for tissue biopsy in recent years (10-12). A companion study of the AURA2 trial reported that patients with EGFRT790M+ plasma, identified by BEAMing ddPCR, may achieve comparable ORRs to osimertinib compared to those with EGFRT790M+ tumor tissues (13), suggesting that liquid biopsy might be a reliable substitute for tumor rebiopsy. Certain patients with EGFRT790M+ plasma are unresponsive to osimertinib therapy possibly due to a low allelic frequency (AF) of plasma EGFR T790M (13). However, the abundance of T790M does not appear to be a predictive factor of the response to osimertinib (14), as it is also significantly associated with circulating tumor DNA (ctDNA) levels which vary markedly among individuals and is influenced by multiple factors (15). Therefore, osimertinib treatment response may be correlated with the relative allelic frequency (RAF) of EGFR T790M, calculated as the ratio of the AF of T790M to the AF of the sensitizing mutation (16). Previous studies have shown that multiple resistant subclones, in addition to T790M, can arise simultaneously following EGFR-TKI exposure in approximately 50% of patients in whom targeting T790M alone is ineffective (10,11). Some studies have proposed the RAF of T790M as a predictor of patient response to osimertinib or another 3rd generation TKI, rociletinib (17-19). A lower ratio of the allele fraction for EGFR T790M to activating EGFR mutations was reported in NSCLC that associated with lower efficacy of osimertinib in subsequent lines of treatment using tissues (20). However, these studies did not elucidate which subgroup of patients with T790M positive plasma are less responsive to 3rd generation TKIs, and what is the concurrent resistance mechanisms.
In this study, we investigated which subgroup of patients, classified by T790M RAF, might have concurrent resistance mechanisms in addition to T790M and poor response to osimertinib. A cutoff value of T790M RAF was established for the prediction of the coexistence of T790M and additional alterations in predicting response to osimertinib, highlighting NGS-determined plasma T790M RAF as a potential prognostic predictor for patients with progressive NSCLC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-915).
Methods
Patients and study design
This study was approved by the Institutional Review Board of West China hospital, Sichuan University, Chengdu, China. A written informed consent was provided by each patient. This study was registered on http://www.chictr.org.cn (registration number: ChiCTR-DDD-16007900). All procedures performed in studies involving human participants were in accordance with the Helsinki declaration (as revised in 2013).
Two independent cohorts of patients with NSCLC, a discovery cohort (n=32) and a validation cohort (n=21), admitted to West China Hospital from August 2015 to April 2017 were included in this study. In the discovery cohort, patients with progressive NSCLC post EGFR-TKI resistance were included if both the repeat biopsy tissue and blood samples were obtained concurrently. The genomic profiles derived from tissue (conventional methods and/or NGS) and plasma (NGS) were compared to find which subgroup of patients carried concurrent resistance mechanisms in addition to T790M. In the validation cohort, patients resistant to first-line EGFR-TKIs were included if they were positive for T790M, determined using an NGS-based plasma assay, and treated with osimertinib. We tested whether patients with concurrent resistance mechanisms and T790M RAF had poor response rates to osimertinib.
Tumor tissue DNA extraction and driver gene testing
DNA was extracted from tissue samples using a QIAamp DNA FFPE tissue kit (Qiagen, CA, USA) according to the manufacturer’s instructions. DNA concentration was measured using a Qubit dsDNA assay kit (Life Technologies, Carlsbad, CA, USA). ARMS-PCR was performed to detect EGFR-sensitizing mutations and T790M. This was carried out using Amplification Refractory Mutation System technology as the ADx EGFR Mutations Detection Kit (Amoy Diagnostics, China) had been approved for clinical application by the State Food and Drug Administration in China. Fluorescence in situ hybridization (FISH) was performed to assess EGFR amplification and MET amplification as previously described (20-23).
Preparation of plasma cell-free DNA (cfDNA)
Whole blood (10 mL) was collected in K3EDTA-containing and cfDNA blood collection tubes (Streck, La Vista, Nebraska, USA) and centrifuged at 2,000 g for 10 minutes at 4 °C within 72 hours after collection. The supernatant was transferred to a fresh 15 mL centrifuge tube without disturbing the buffy coat layer, and was subjected to an additional centrifugation at 16,000 g for 10 minutes at 4 °C. The supernatant was then transferred to a new tube and stored at −80 °C until further analysis. Circulating cfDNA was extracted from the plasma samples using a QIAamp circulating nucleic acid kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. Quantification of cfDNA was performed using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, California, USA) and dsDNA HS assay kits (Life Technologies).
NGS library preparation and capture-based targeted DNA sequencing
DNA shearing was performed using a Covaris M220, followed by end repair, phosphorylation and adaptor ligation. DNA concentration and genomic DNA quality were measured using a Qubit dsDNA assay and 260 nm/280 nm absorption ratios, respectively. Fragments of size 200–400 bp were selected using Agencourt AMPure XP beads (Beckman Coulter, CA, USA) followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. A bioanalyzer high-sensitivity DNA assay was then performed to assess the quality and size of the fragments, and indexed samples were sequenced on a Nextseq500 sequencer (Illumina, CA, USA) with pair-end reads.
Sequence data analysis
Sequence data were mapped to the human genome (hg19) using the BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan, respectively. Plasma samples were compared against their own white blood cells to identify somatic variants. Variants were filtered using the VarScan fpfilter pipeline, and loci with depth less than 100 were filtered out. At least 2 and 5 supporting reads were needed for INDELs in plasma and tissue samples, respectively, while 8 supporting reads were needed for SNVs to be called in both plasma and tissue samples. According to the ExAC, 1,000 Genomes, dbSNP, ESP6500SI-V2 databases, variants with population frequency over 0.1% were grouped as SNPs and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translocation analysis was performed using Tophat2 and Factera.
Statistical analysis
Continuous data were presented as mean ± standard deviation (normal distribution) or median, range (skewed distribution), while categorical data were presented as frequency and percentage. Comparisons of response rates and disease control rates (DCRs) were conducted using the continuity correction method. Progression free survival rates were estimated using the Kaplan-Meier method. A value of P<0.05 was considered statistically significant. Statistical analysis was carried out using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA).
Results
Patient characteristics
The demographic and clinical characteristics of the two cohorts are shown in Table 1. In the discovery cohort, 32 patients (12 males and 20 females; mean age = 58.5 years) harboring EGFR sensitizing mutations 19del (n=14) or L858R (n=18) with both re-biopsy tumor and ctDNA genomic profiling were included. All patients were treated with the first-generation TKIs gefitinib (n=22), erlotinib (n=3), or icotinib (n=7). The median PFS of the 1st generation TKIs in this cohort was 9 months. Among the discovery cohort, 14 patients had symptomatic progression and the remaining 18 patients had asymptomatic progression. In the validation cohort, 21 patients (5 males and 16 females; mean age =60.4 years) harboring 19del (n=10) or L858R (n=11) and resistant to TKIs were included. All of them had T790M positive plasma as determined by NGS, and were treated with osimertinib at the time of progression, and 12 patients were subjected to re-biopsy. The median PFS of 1st generation TKIs in this cohort was 13 months.
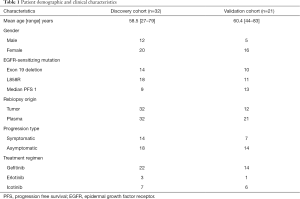
Full table
Concordance and heterogeneity observed in the mutation spectrum between tissue and plasma
To evaluate the sensitivity and reliability of genomic mutation detection, we performed ARMS-PCR for EGFR mutations on re-biopsy tissue samples from 32 TKI-treated progressive NSCLC patients, and performed capture-based targeted deep-sequencing on matched plasma and tissue samples obtained from 30 patients. Two patients (P22 and P31) who had inadequate tissue samples for NGS were excluded. EGFR and MET amplifications were successfully evaluated by FISH in 28 patients (Figure S1). For EGFR sensitizing mutations and T790M in tissue samples, the concordance rates between ARMS-PCR and NGS were 100% and 87%, respectively. For the 4 discordant tissue samples, NGS detected T790M that was missed by ARMS-PCR. The concordance between tissue and plasma was 93.8% (30/32) for EGFR sensitizing mutations compared to only 62.5% (20/32) for T790M. The detection rates of T790M from ctDNA and tissue biopsy were 53.1% (17/32) and 40.6% (13/32), respectively (Figure 1). These findings suggest that NGS may be more sensitive than ARMS-PCR in capturing T790M in tissue samples, and might detect additional T790M+ cases in plasma that are missing in tissue samples.
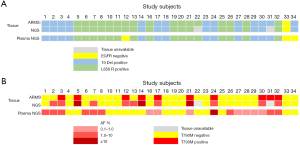
In addition, 130 genetic aberrations involving 27 genes were identified from 30 tissue biopsy samples which were adequate for NGS (Figure S2A). In these aberrations, MET amplification, another resistance mechanism to EGFR-TKI, was observed in 25.8% (8/31) patients in combination with other mutations (Figure S2A). Other frequently occurring mutations included, but were not limited to, TP53, RB1, and PTEN in 59%, 19% and 13% patients, respectively. Furthermore, 135 genetic variants involving 36 genes were identified in plasma samples (Figure S2B). Using the mutation spectrum derived from tissue samples as a reference, we achieved 78% sensitivity excluding CNVs.
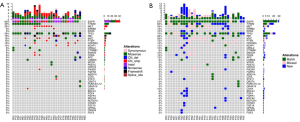
Patients with a low RAF of EGFR T790M are more likely to harbor concomitant resistance mechanisms
In the discovery cohort, 59% patients (20/34) harbored EGFR T790M, of whom 60% (12/20) harbored additional resistance mechanisms (Figure 2A). As shown in Figure 2B, the resistance mechanisms in 25 patients (81%) were identified, whereas those in the remaining 19% patients (6/31) were unidentified. EGFR T790M was found in the majority of patients (20/31) either as a solo resistance mechanism or in combination with others. MET and ERBB2 amplification were the other two mechanisms that existed as solo resistance mechanisms. In total, 42% (13/31) of patients had solo resistance mechanisms, including EGFR T790M (n=9), MET amplification (n=3) and ERBB2 amplification (n=1), whereas 39% (12/31) of patients harbored concomitant resistance mechanisms. We next sought to investigate the association between T790M and the presence of concomitant resistance mechanisms. The RAF (ranging from 1.3% to 100%) was plotted in descending order along with concomitant resistance mechanisms. As shown in Figure 2A, 12 patients harbored concomitant resistance mechanisms in addition to EGFR T790M, among whom 9 had an RAF less than 20%.
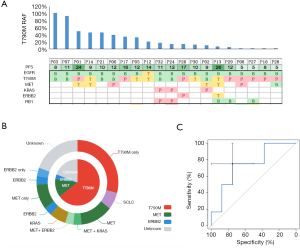
To evaluate whether 20% T790M RAF may serve as an optimal cutoff in distinguishing populations who are more likely to harbor concomitant resistance mechanisms, we conducted a receiver operating characteristic (ROC) curve analysis of the presence of concomitant resistance mechanisms against EGFR T790M RAF. As shown in Figure 2C, the area under the curve was 74%. Using 20% as a cutoff, patients harboring low RAF (<20%) were more likely to have concomitant resistance mechanisms in addition to T790M (P=0.018), suggesting that RAF (<20%) can be used as a marker to distinguish populations carrying additional genetic alterations other than T790M, and theoretically, having poorer response rates to osimertinib.
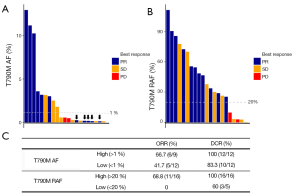
T790M RAF predicted poorer osimertinib response
To explore whether T790M RAF can potentially predict osimertinib response, we recruited an independent cohort consisting of 21 EGFR T790M-positive NSCLC patients who experienced progression from previous 1st generation EGFR-TKI therapy and received osimertinib treatment. Among these patients, 11 experienced partial response (PR), 8 stable disease (SD) and 2 progressive disease (PD). Figure 3A demonstrates that absolute T790M AF is not an ideal marker to predict osimertinib efficacy. Patients with T790M AF less than 1% achieved an ORR of 41.5% and a DCR of 83.3%, with no significant differences compared to patients with T790M AF higher than 1%. Using T790M RAFs as a biomarker, Figure 3B shows that patients with low T790M RAF achieved 0% ORR and only 60% DCR, significantly lower than those with high T790M RAF (P=0.012 and P=0.048, respectively), suggesting that low T790M RAF is likely a contraindication for osimertinib treatment. The ORR and DCR of osimertinib for patients classified by T790M AF and RAF are listed in Figure 3C.
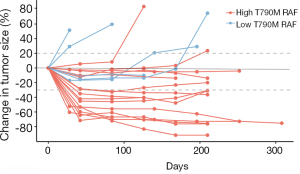
We further evaluated tumor size changes in this cohort of patients. As shown in Figure 4, among the 16 patients with T790M RAF greater than 20%, 14 of them did not experience tumor growth during treatment. Furthermore, tumor size reductions were observed in all patients except 1 at certain points during treatment, suggesting a sensitive response to osimertinib in this group. Contrastingly, among the 5 patients with T790M RAF less than 20%, 2 exhibited continuous tumor growth and 3 experienced a short period of tumor reduction followed by steady tumor growth, which indicates a poor response to osimertinib in this group. These findings further support the positive correlation between T790M RAF and osimertinib response.
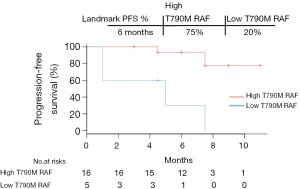
Low T790M RAF indicated poor PFS with osimertinib treatment
We next sought to investigate the correlation between T790M RAF and PFS in the validation cohort by performing a Kaplan-Meier survival analysis. As shown in Figure 5, disease progression occurred in all 5 patients with low T790M RAF with a median PFS of 5 months. The median PFS of patients with high T790M RAF was not reached at the end of follow-up, which was markedly longer than the median PFS of patients with low RAF (P=0.003, HR =11.37, 95% CI: 2.28–56.59). Taken together, these data suggest that low T790M RAF is an indicator of poor osimertinib efficacy in this population of patients.
Discussion
In the present study, we demonstrated that NGS of liquid biopsy samples can effectively detect the mutation spectrum of solid tumors, providing an alternative approach to tissue-based gene profiling in revealing the mechanisms underlying drug resistance to NSCLC. Our results showed that genotyping in matched rebiopsy tissue and plasma samples achieved 93.8% concordance for EGFR sensitizing mutations and 62.5% for T790M. In the discovery cohort, patients with low plasma T790M RAFs (<20%) were more likely to carry concurrent resistance mechanisms. In the independent validation cohort, patients with low plasma T790M RAFs exhibited a poorer response to osimertinib than those with high plasma T790M, suggesting that unresponsiveness to osimertinib may be due to other resistance mechanisms in addition to T790M. Thus, T790M RAF may represent a prognostic stratification in addition to the current binary assessment of T790M status (positive vs. negative) for patients treated with 3rd generation TKIs (24).
NGS-based liquid biopsy is a well-established alternative approach to tissue rebiopsy (10-12). In our study, the results of NGS-detected EGFR sensitizing mutations between matched tissue and plasma samples were highly concordant (93.8%). The by-variant sensitivity was 78% excluding CNVs and the by-patient sensitivity was 93.5%, consistent with previous studies (25,26). This suggests that NGS-based liquid biopsy is a reliable approach for the identification of genetic alterations. In addition, the concordance rate for NGS-detected T790M between tissue and plasma samples was 62.5% because more T790M cases were captured by NGS in plasma samples than in tissues samples. This suggests that NGS-based liquid biopsy is a highly sensitive method for T790M detection, especially when tissue samples are unavailable. Although EGFR T790M is responsible for over 50% of acquired resistance to first-generation EGFR-TKIs (4,27,28), other genes, such as MET, KRAS, ERBB2, and RB1 (20-22,29,30), have been found to be altered simultaneously with T790M, contributing to the underlying mechanism of TKI resistance. Our data showed that 60% of patients harboring T790M carried other resistance mechanisms concurrently, which is partially consistent with previous studies (31-33). Based on the high sensitivity and reliability of NGS in detecting EGFR sensitizing mutations as well as T790M in plasma, RAF of plasma EGFR T790M, could be used to differentiate subgroups of patients harboring different levels of T790M. By analyzing the T790M RAFs of patients who carried solo T790M or in combination with other resistant genomic alterations, we found 20% was an optimal cutoff to distinguish these two subgroups. Patients in the discovery cohort with low plasma T790M (<20%) were more likely to carry concurrent resistance mechanisms in addition to T790M. This cutoff value was further tested in an independent cohort receiving osimertinib therapy. The results showed that patients with low plasma T790M RAF (<20%) had poorer ORR and PFS than those with high RAF (≥20%). These data collectively suggest that a poor response to osimertinib in the patients with low plasma T790M RAF is possibly due to the cooperation of multiple resistance mechanisms in these patients. This finding is consistent with several previous studies showing that quantification of T790M mutant alleles can be used to predict outcome after TKI treatment using 50% as a cutoff value (17-19). However, based on our results, we suggest that patients with T790M RAF between 20–50% should still be prescribed osimertinib as some patients could also benefit from osimertinib treatment and achieve PR or SD. Thus, the cutoff value of 50% may be too high to identify all the patients responding positively to osimertinib therapy. Since our study was conducted using a small sample size, the feasibility of a 20% cutoff should be tested in a larger population in future studies.
Clinical studies have shown that osimertinib treatment may achieve approximately 60% ORR in T790M-positive patients (34,35). Patients who do not respond to osimertinib treatment may harbor other resistance mechanisms in addition to T790M, and the percentage of T790M-positive tumor cells is positively correlated with the response to osimertinib (14-16,34). Our results suggest that NGS of plasma may serve as an effective approach to quantify the percentage of T790M-positive tumor cells in order to predict the response to osimertinib. T790M RAF in plasma may better reflect the proportion of T790M-positive tumor cells compared to rebiopsy tissue due to tumoral heterogeneity. Therefore, NGS of plasma should also be performed even when tissue biopsy is available, and T790M RAF should be taken into consideration before commencing osimertinib treatment. In addition, plasma-based NGS for detection of MET amplification and RB1 mutations may provide information associated with innate and acquired resistance mechanisms. However, due to the limited sample size in this study, a prospective study involving more patients is needed.
In summary, our results suggest that NGS-detected plasma T790M RAF with an optimal cutoff value of 20% is useful for distinguishing subgroups of patients resistant to 1st generation EGFR-TKIs. Patients with low T790M RAF (<20%) are more likely to carry concurrent resistance mechanisms and are less likely to benefit from osimertinib treatment. T790M RAF determination should be considered a potential standard procedure in NSCLC treatment decisions.
Acknowledgments
The author would thank Han Han-Zhang and Junyi Ye for revision of the manuscript.
Funding: This work was supported by the Transformation Projects of Sci-Tech Achievements of Sichuan Province (2016CZYD0001), the Sci-Tech Support Program of the Science and Technology Department of Sichuan Province (2016SZ0073), the National Major Sci-Tech Project (2017ZX10103004-012) and the National Key Development Plan for Precision Medicine Research (2017YFC0910004).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-915
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-915
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-915). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the Helsinki declaration (as revised in 2013). This study was approved by the Institutional Review Board of West China hospital, Sichuan University, Chengdu, China. A written informed consent was provided by each patient. This study was registered in http://www.chictr.org.cn (register number: ChiCTR-DDD-16007900).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Chen D, Chu T, Chang Q, et al. The relationship between preliminary efficacy and prognosis after first-line EGFR tyrosine kinase inhibitor (EGFR-TKI) treatment of advanced non-small cell lung cancer. Ann Transl Med 2019;7:195. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [Crossref] [PubMed]
- Ariyasu R, Nishikawa S, Uchibori K, et al. High ratio of T790M to EGFR activating mutations correlate with the osimertinib response in non-small-cell lung cancer. Lung Cancer 2018;117:1-6. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Stockley T, Souza CA, Cheema PK, et al. Evidence-based best practices for EGFR T790M testing in lung cancer in Canada. Curr Oncol 2018;25:163-9. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Belchis DA, Tseng LH, Gniadek T, et al. Heterogeneity of resistance mutations detectable by nextgeneration sequencing in TKI-treated lung adenocarcinoma. Oncotarget 2016;7:45237-48. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014;14:294. [Crossref] [PubMed]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Yang X, Zhuo M, Ye X, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 2016;7:20810-24. [Crossref] [PubMed]
- Passiglia F, Rizzo S, Maio MD, et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis. Sci Rep 2018;8:13379. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Marchetti A, Palma JF, Felicioni L, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 2015;10:1437-43. [Crossref] [PubMed]
- Tian P, Wang Y, Wang W, et al. High-throughput sequencing reveals distinct genetic features and clinical implications of NSCLC with de novo and acquired EGFR T790M mutation. Lung Cancer 2018;124:205-10. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Tanaka A, Sueoka-Aragane N, Nakamura T, et al. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer 2012;75:89-94. [Crossref] [PubMed]
- Tsuda H, Akiyama F, Terasaki H, et al. Detection of HER-2/neu (c-erb B-2) DNA amplification in primary breast carcinoma. Interobserver reproducibility and correlation with immunohistochemical HER-2 overexpression. Cancer 2001;92:2965-74. [Crossref] [PubMed]
- Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 2009;27:1667-74. [Crossref] [PubMed]
- Varella-Garcia M, Diebold J, Eberhard DA, et al. EGFR fluorescence in situ hybridisation assay: guidelines for application to non-small-cell lung cancer. J Clin Pathol 2009;62:970-7. [Crossref] [PubMed]
- Ichihara E, Lovly CM. Shades of T790M: Intratumor Heterogeneity in EGFR-Mutant Lung Cancer. Cancer Discov 2015;5:694-6. [Crossref] [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [Crossref] [PubMed]
- Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2008;14:2895-9. [Crossref] [PubMed]
- Del Re M, Tiseo M, Bordi P, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget 2017;8:13611-9. [Crossref] [PubMed]
- Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377. [Crossref] [PubMed]
- Chen LY, Molina-Vila MA, Ruan SY, et al. Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: A systematic review and meta-analysis. Lung Cancer 2016;94:46-53. [Crossref] [PubMed]
- Ko R, Kenmotsu H, Serizawa M, et al. Frequency of EGFR T790M mutation and multimutational profiles of rebiopsy samples from non-small cell lung cancer developing acquired resistance to EGFR tyrosine kinase inhibitors in Japanese patients. BMC Cancer 2016;16:864. [Crossref] [PubMed]
- Kato Y, Hosomi Y, Watanabe K, et al. Impact of clinical features on the efficacy of osimertinib therapy in patients with T790M-positive non-small cell lung cancer and acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors. J Thorac Dis 2019;11:2350-60. [Crossref] [PubMed]
- Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in Pretreated T790M-Positive Advanced Non-Small-Cell Lung Cancer: AURA Study Phase II Extension Component. J Clin Oncol 2017;35:1288-96. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


