
Investigation on the potential of circulating tumor DNA methylation patterns as prognostic biomarkers for lung squamous cell carcinoma
Introduction
Lung cancer remains the leading cause of cancer-related death worldwide, including China (1-4). Among the non-small cell lung cancer (NSCLC) histological subtypes, lung squamous cell carcinoma (LUSC) is one of the most prevalent, accounting for approximately 15–30% of all lung cancers diagnosed in China (1,5-9). Histologically, LUSC is an epithelial malignancy characterized by keratinization and/or intercellular bridges and demonstrates positivity for squamous differentiation markers using immunohistochemistry (7). LUSC is strongly associated with exposure to tobacco and is predominantly diagnosed in males (1,8-10). The 5-year survival rate of LUSC is approximately 18% (1), with survival outcomes remaining unchanged over the last decade. The tumor-node-metastasis (TNM) staging system remains as the primary guide for the treatment and prognostication for NSCLC, including LUSC (11,12). As the standard of care, patients with early-stage LUSC are managed with surgery and adjuvant chemotherapy, with or without radiotherapy (13). Meanwhile, patients with advanced-stage LUSC are treated with cytotoxic chemotherapy as a first-line treatment, particularly with platinum-based doublet, and are recommended to undergo molecular testing for actionable mutations (13). However, due to the low frequency of actionable mutations typically found in lung adenocarcinoma, such as EGFR and ALK (14-16), the use of targeted therapy remains limited among patients with advanced LUSC. Late diagnosis and the lack of actionable mutations are major contributors to the poor prognosis of LUSC. Numerous efforts have been invested in identifying molecular factors with either prognostic or predictive value in LUSC; however, no biomarker has been validated for use in standard clinical practice (10,17,18). Thus, there is an urgent need to identify prognostic and predictive biomarkers to improve outcomes of patients with LUSC.
DNA methylation is a major epigenetic modification by which methyl groups are added to critical regions of DNA for the precise control of gene expression. The key biological processes under epigenetic control include the cell cycle, DNA repair, cell proliferation, and apoptosis, which are critical pathways involved in tumor development and progression (19,20). Aberrant DNA methylation, including genome-wide hypomethylation of repetitive elements and CpG-poor regions, and regional hypermethylation of CpG islands at gene promoters, has been implicated in the early stages of carcinogenesis in various solid tumors including LUSC (18,20-27). Numerous studies have explored the potential of epigenetic deregulation as a biomarker of early detection, cancer surveillance/disease monitoring, and prognosis of various human cancers (26,28-30).
Liquid biopsy samples are now being used as minimally-invasive alternative sources of tumor DNA for next-generation sequencing (NGS)-based genomic testing (31-34). The advances in molecular detection using liquid biopsy samples are particularly advantageous for patients who have tumors that are inaccessible or have a limited volume of tumor biopsy specimen. The presence of circulating tumor DNA (ctDNA) derived from blood samples, the most commonly used liquid biopsy specimen, is associated with minimal residual disease from resected early-stage lung cancer and serves as an early biomarker for cancer recurrence (29,34). With the possibility of longitudinal sampling, considerable attention has been invested towards exploring the utility of ctDNA methylation profiling for the early detection of cancer recurrence or relapse (30,33,35-43). Numerous genes have been identified to be differentially methylated in blood samples of patients with early-stage lung cancer, including SHOX2, RASSF1A, and p16, which could potentially serve as predictive biomarkers (30,33,35-42). As compared to lung adenocarcinoma, DNA hypermethylation of SHOX2 was highest in small cell and LUSC tumor tissue samples, indicating that methylation status is distinct among lung cancer histologies (40). However, investigations on ctDNA methylation patterns that enable the prediction of prognosis at the time of diagnosis were performed on lung cancer patients in general and independent investigation on LUSC is very limited. In our study, we aimed to identify differential methylation signatures in certain genomic CpG regions from blood samples at initial diagnosis that are associated with survival, and potentially predict survival outcomes in Chinese patients with LUSC. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1070).
Methods
Patients
A total of 26 consecutive patients with stage IA–IV lung cancer with confirmed squamous cell histology diagnosed at our institution between March 2017 and September 2019 were prospectively included in this study. Clinical staging and histology were evaluated according to the eighth edition of the American Joint Committee on Cancer (11) and the 2015 World Health Organization classification of lung tumors (7), respectively. The main study inclusion criteria are as follows: (I) pathologically-confirmed squamous cell carcinoma; (II) provided consent to submit blood samples at diagnosis; (III) provided written informed consent to participate in the study. The main exclusion criteria are as follows: (I) non-squamous cell lung cancer histology; (II) history of other malignant tumors within the last 5 years; (III) failure to obtain blood samples prior to receiving treatment; (IV) refusal or withdrawal of consent. Baseline clinicopathological information was collected, including age at diagnosis, gender, smoking status, and treatment information, and is summarized in Table S1. The cohort was comprised of a majority of males (88.5%; 23/26), with a median age of 63 years (range, 38–78 years). All patients received therapeutic management based on their clinical stages following the guidelines of the National Comprehensive Cancer Network and the decision of the physician-in-charge. Baseline blood samples were obtained from each patient at the time of initial visit/diagnosis. The study was approved by our Institutional Review Board (approval number: NCC1576) and performed in compliance with the ethical standards of the institutional and/or national research committee. This study was also carried out in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided written informed consent before inclusion in the study.
DNA isolation
DNA isolation and subsequent NGS library preparation were performed at Burning Rock Biotech, a College of American Pathologists (CAP)-accredited and Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratory. In brief, circulating cell-free DNA (cfDNA) was isolated from plasma samples using QIAamp Circulating Nucleic Acid kit, according to the manufacturer’s standard protocol (Qiagen, Hilden, Germany). cfDNA quantity and quality were estimated using the Bioanalyzer 2100 instrument and high sensitivity DNA assay kit (Agilent Technologies, CA, USA). A minimum of 50 ng DNA was required for the NGS library construction.
Unique molecular identifier (UMI)-based targeted sequencing and data analysis
Targeted sequencing was performed as described previously (44). The cfDNA fragments were ligated with adapters containing UMIs. Target capture was performed using a panel consisting of 168 cancer-related genes (Burning Rock Biotech), spanning 273 kilobases of the human genome. The UMI-tagged indexed samples were subsequently sequenced on an Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) using 2×150 base pair (bp) cycles and a target sequencing depth of 30,000×. Sequencing data were analyzed using bioinformatics pipelines optimized to accurately distinguish between somatic mutations and sequencing artifacts.
Bisulfite targeted sequencing and data analysis
Whole-genome bisulfite sequencing library preparation was performed using the brELSA (Burning Rock Biotech) as described previously (35). Briefly, purified cfDNA were treated with sodium bisulfite (EZ-96 DNA Methylation-Lightning MagPrep, Zymo Research, Orange, CA, USA). The converted fragments underwent two series of ligation with appropriate adapters, amplification, and purification to obtain the whole-genome bisulfite sequencing library. Target enrichment was performed using custom-designed lung-cancer methylation profiling RNA baits covering 80,672 CpG sites, spanning 1.05 megabases of the human genome. The target libraries were finally quantified by real-time PCR (Kapa Biosciences, Wilmington, MA, USA) and sequenced on a NovaSeq 6000 (Illumina, San Diego, CA, USA) using 2×150 bp cycles. Bisulfite sequencing data analysis was performed using an optimized pipeline. Custom adapter sequences and low-quality bases were removed using Trimmomatic (v.0.32). Sequence alignment of paired-end reads to CtoT- and GtoA-transformed hg19 genome was performed using BWA-meth (v.0.2.2) (45). PCR duplicates from aligned sequences were marked with Samblaster (v.0.1.20) (46). Sambamba (v.0.4.7) (47) was then used to remove the sequencing reads with either low mapping quality (MAPQ <20) or improper pairing. To eliminate potential double-counting of methylation calls, overlapping reads from paired-read sequences were clipped and merged using in-house scripts.
Analysis of methylation patterns
Methylation levels in this study were analyzed and represented as scores to reflect methylation features of the sample. Using the methods described previously (35), the 80,672 CpG sites included in the panel were grouped into 8,312 methylation blocks to account for linkage disequilibrium. Methylation blocks were defined as the genomic region between the neighboring CpG sites with the r2 value calculated based on our modified correlation matrix. In brief, Pearson’s correlation analysis was performed to calculate the difference among the methylation frequencies of each pair of CpG sites, normalized by the difference in genomic distance and methylation level. Methylation frequencies for the entire set of methylation blocks were calculated as the ratio between the total number of Cs at all CpG sites within a methylation block, and the total number of C+Ts at the same methylation block.
Methylation pattern value (MPV1) was defined as the differential methylation pattern of CpG sites in a certain genomic region with an interval of 0–1 within a methylation block, and is represented by the following formula in Eq. [1]:
[1]
Wherein: n refers to the number of sequencing reads for the multiple CpG sites that cover the methylation block; Li refers to the number of CpG sites within the methylation block that is covered by the i-th sequencing read; mi refers to the number of methylation blocks with consecutively methylated CpG sites which appeared in the i-th sequencing read; lij indicates the number of methylated CpG sites per methylation block.
Since each of the 8,312 methylation blocks had their individual MPV1 score, a methylation score was also calculated per patient sample using Eq. [2].
[2]
Wherein: n refers to the number of methylated blocks; Ti refers to the value corresponding to the i-th methylation block estimated for the patient circulating tumor DNA (ctDNA) sample; Hi refers to the value corresponding to the i-th methylation block estimated for a group of asymptomatic healthy samples, methylation data were derived from ctDNA mutation profiling of 198 asymptomatic healthy patients; N indicates the number of asymptomatic healthy patients included in the analysis; Hij refers to the value corresponding to the i-th methylation block for j-th sample from the asymptomatic healthy patients;
Statistical analysis
Statistical analysis was performed using the R statistics package (R version 3.4.0; R: The R-Project for Statistical Computing, Vienna, Austria). Statistical significance was defined as P values <0.05. Differences between groups were calculated using a two-tailed Student’s t-test, as appropriate. Overall survival (OS) was defined from the date of diagnosis until death or the last day of follow-up. OS values corresponding to the last day of follow-up for patients who are still alive were censored and were indicated by tick marks in the survival curves. The data cut-off was October 1, 2019 with a median follow-up of 6 months (ranging from 1 month to 34 months). Survival differences between patients with certain maximum allelic fraction (maxAF) percentages were estimated using the Kaplan-Meier method and compared using the log-rank test. Univariate Cox proportional-hazards model was used to estimate the correlation between molecular features and survival outcome. Least absolute shrinkage and selection operator (LASSO) was implemented using the glmnet package to perform a regression analysis for identifying the smallest possible methylation block set that correlated with maxAF.
Results
Molecular profile of the cohort
To understand the baseline somatic mutation profile of the patients, targeted sequencing analysis was performed using blood samples obtained at the initial visit. Among the 26 patients with adequate baseline samples, the mutation detection rate was 80.8% [20/26], with the remaining 6 patients found to be wild-type for genes included in the panel. The most frequently mutated gene in the cohort was TP53, with a mutation rate of 73.0% [19/26]. Among the classic lung cancer driver mutations, PIK3CA (n=3), EGFR amplification (n=2), EGFR exon 19 deletion (n=1), KRAS Q61R (n=1), and MET amplification (n=1) were identified from our cohort. The mutation profile of the cohort is illustrated in Figure S1.
Correlation between maxAF and survival outcome
The maxAF represented the somatic mutation detected from the patient, and was defined as the highest allelic fraction observed among all the mutations detected from their blood sample, regardless of gene or mutation site. The median maxAF of the cohort was 2.06%, ranging between 0 and 52.86%. Among the 26 evaluable patients, 65.5% [17/26] of the patients had a maxAF of ≤5%, while the remaining 9 patients had a maxAF >5%. Patients with a maxAF >5% had significantly shorter OS compared to those with a maxAF ≤5% (P=0.020, Figure 1). The median OS for patients with a maxAF >5% was 7.1 months, while patients with a maxAF ≤5% had a median OS of 54.6 months.
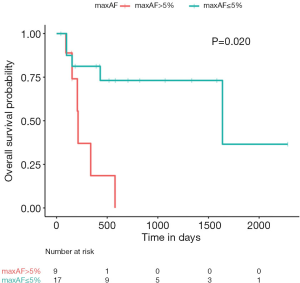
Cox regression analysis further revealed an association between OS outcomes and maxAF [hazard ratio (HR) =54.12; 95% confidence interval (CI): 1.89–1,547.00; P=0.020], suggesting that ctDNA somatic mutations, represented by maxAF, hold potential as prognostic biomarkers for LUSC.
Correlation between methylation features and survival outcomes
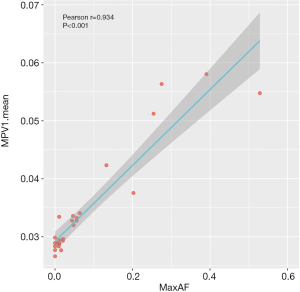
We further analyzed the methylation profile of our cohort at the time of diagnosis to understand their epigenetic status at baseline. A strong correlation was found between maxAF and differential methylation patterns expressed as MPV1 (Pearson’s r=0.934, P<0.001; Figure 2). Consistently, Cox regression analysis also revealed an association between OS and differential methylation patterns (HR =2.377; 95% CI: 1.283–4.405; P=0.006), indicating that the higher the methylation levels, the worse the survival outcomes.
Interestingly, in-house methylation data derived from 251 LUSC patient tissue samples at various stages (stage I–IV) revealed that a total of 402 methylation blocks correlated with OS. When considering only these 402 methylation blocks, the ctDNA methylation patterns of our cohort at baseline, expressed as MPV1, also demonstrated a strong correlation with OS (P=0.027, adjusted with maxAF, smoking, and stage).
These data suggest that ctDNA methylation status at the time of diagnosis could serve as a potential prognostic biomarker for LUSC.
Identifying maxAF-related methylation signatures
We further used LASSO to identify certain methylation blocks that were associated with maxAF and OS. A total of 1,956 methylation blocks were found to be differentially methylated from 21 samples with maxAF >0%, and were significantly correlated with maxAF (P<0.05). Using 7-fold cross-validation, the lambda.min and lambda.1se of the generalized linear model were used to determine the optimal methylation signature that was correlated with maxAF. Figure 3 illustrates the lambda and the corresponding mean-squared error (MSE) for various methylation block features generated from the LASSO model.
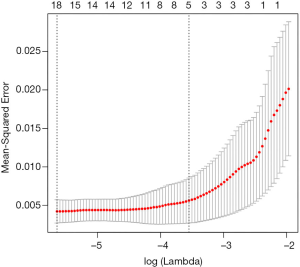
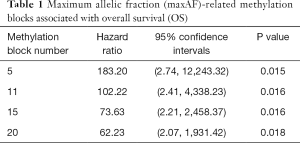
When the lambda of the model equalled lambda.min yielding the minimum average cross-validated error of 3.5E-03, the model which included five methylation blocks had an MSE of 4.2E-03 (95% CI: 2.7E-03, 5.7E-3). When the lambda equalled lambda.1se, yielding the most normalized model whereby the error within 1 standard error of the minimum was 2.9E-02, the model which included 20 methylation blocks had an MSE of 5.6E-03 (95% CI: 2.7E-03, 8.5E-3). Using the model constructed with five methylation blocks on all of the 26 patient samples from the cohort including those with maxAF=0, the predicted methylation signature value was significantly associated with OS (P=0.015), and the corresponding HR was the largest compared to models constructed with 11, 15, and 20 methylation blocks (Table 1). Table 2 lists the genomic position of the five differentially methylated blocks associated with prognosis. Table S2 lists the genomic position of all the methylation blocks identified by LASSO.
Full table

Full table
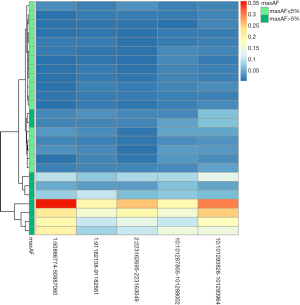
Interestingly, MPV1 corresponding to the five methylation blocks was higher among the patients with maxAF >5% compared to those with maxAF ≤5% (Figure 4). Table S3 summarizes the clinicopathological features, maxAF, and MPV1 scores for the 5 methylation blocks of the cohort. Patients with higher MPV1 and maxAF >5% had shorter OS, indicating that these differential methylation signatures from the five methylation blocks could serve as an alternative to maxAF, and potentially serve as prognostic biomarkers for LUSC.
Discussion
In addition to the clinical value of somatic mutation profiling in guiding therapeutic management for patients with lung cancer, mounting evidence supports the utility of DNA methylation signatures as diagnostic, prognostic, or predictive biomarkers (29-34). Compared to invasive tissue sampling, blood samples can be obtained in a minimally-invasive manner and sampled repeatedly during the treatment course. CtDNA, derived from DNA fragments shed into the plasma via necrosis, apoptosis, or active release of DNA from tumor cells, can reflect dynamic changes in the genetic and epigenetic status of the tumor without the intra-tumor heterogeneity associated with tissue biopsies (31-34). The concentration of ctDNA from the plasma-derived cfDNA is highly dependent on the tumor burden. Therefore, the detection of somatic mutations from ctDNA is severely limited in early-stage tumors (44). As compared with somatic mutations, DNA methylation in promoters of various tumor suppressor genes is more ubiquitous and occurs at the early stages of lung tumor development (18,20-27), which improves the sensitivity of detecting aberrant DNA methylation from cfDNA. Numerous reports have identified the promoter region of a single gene or a panel of genes that are differentially methylated in NSCLC; however, histology-based subcohort analysis on LUSC is limited (30,33,35-42). As compared to lung adenocarcinoma, LUSC has shown a histologically distinct methylation pattern in certain gene promoters (40); however, no study has yet reported on specific prognostic markers in LUSC.
In this study, we performed parallel somatic mutation and methylation profiling of ctDNA obtained from Chinese patients with various stages of LUSC to identify the molecular factors that can predict survival outcomes at the time of diagnosis. Our data demonstrated that maxAF >5%, derived from ctDNA somatic mutation profiling at baseline, was significantly correlated with shorter OS. Furthermore, we identified five genomic regions or genes that were differentially methylated at baseline and were significantly correlated with OS, suggesting their potential to predict prognosis even at the time of diagnosis. To the best of our knowledge, this study is the first to identify methylation signatures from the ctDNA of patients with LUSC that are significantly correlated with their prognosis. These methylation signatures are promising biomarkers for predicting survival outcome at the time of diagnosis for this subset of patients. Our findings highlight the potential clinical utility of ctDNA somatic mutation and methylation profiling at baseline for predicting the survival outcomes of patients diagnosed with various stages of LUSC. DNA methylation profiling of plasma samples could provide a minimally-invasive alternative to tissue biopsy procedures in the prognostication of patients with LUSC.
The ctDNA somatic mutation profile of our cohort was consistent with the mutation profile of LUSC patients in previous studies (14-16). TP53 was the most commonly mutated gene from our cohort, with an overall mutation rate of 73%. Only 8 patients from our cohort had potentially actionable mutations with available targeted agents. Stratifying the patients into two groups based on maxAF at baseline revealed a strong correlation between maxAF and OS.
ctDNA methylation profiling of our cohort revealed differential methylation patterns in various genomic regions, with 1,956 differentially methylated blocks at baseline correlated with maxAF. Among the 5 differentially methylated regions associated with maxAF and OS, DMRTA2 was also identified to be differentially methylated in lung tumors compared with normal lung samples and benign tumors (25). Moreover, PAX3 (22) and BARHL2 (23) were also identified as hypermethylated from primary LUSC tumors. However, no study has associated the differential methylation of these genes or other genomic regions with the prognosis of patients with LUSC.
Our study is limited by the inclusion of a small cohort of patients recruited from a single center, which might have introduced sampling bias. A prospective cohort study with a larger sample size in a multi-center setting is required to further establish the potential of differential methylation signatures from these genomic regions as prognostic biomarkers of patients with LUSC. Experimental studies on the individual genes that are correlated with prognosis would also advance the understanding of their regulation and functional role. It would also be interesting to analyze longitudinal ctDNA samples from patients with LUSC during their treatment course to establish methylation patterns that could predict treatment response, development of resistance, and disease relapse.
Conclusions
Our study identified molecular features that show promise as prognostic biomarkers for patients with various stages of LUSC at the time of their diagnosis. Our study provides an incremental contribution towards understanding the clinical value of ctDNA methylation and somatic mutation profiling at baseline in predicting the survival outcomes of patients with LUSC.
Acknowledgments
The authors thank all the patients and their families. We also thank the investigators, study coordinators, operation staff, and the whole project team who worked on this study.
Funding: This work is supported by a grant from the Wu Jieping Medical Foundation Precision Medicine for Thoracic Cancer Fund (grant number: 320.6750.17281 to YL). The funding agency had no role in the study design, data collection, analysis, interpretation, manuscript writing, and decision to submit the article for publication.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1070
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1070
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1070). Dr. TH, Dr. AL, Mr. FX, Ms. QK, and Dr. LZ are employed by Burning Rock Biotech. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures involving human participants were performed in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the relevant Institutional Review Board of the Chinese Academy of Medical Sciences & Peking Union Medical College (approval number: NCC1576). Prior written informed consent was obtained from each of the recruited patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cheng TY, Cramb SM, Baade PD, et al. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J Thorac Oncol 2016;11:1653-71. [Crossref] [PubMed]
- Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-iv21. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Zhou F, Zhou C. Lung cancer in never smokers-the East Asian experience. Transl Lung Cancer Res. 2018;7:450-63. [Crossref] [PubMed]
- Fan H, Shao ZY, Xiao YY, et al. Incidence and survival of non-small cell lung cancer in Shanghai: a population-based cohort study. BMJ Open 2015;5:e009419. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Sun X, Wang M, Xu R, et al. Prognostic model based on circular RNA circPDK1 for resected lung squamous cell carcinoma. Transl Lung Cancer Res 2019;8:907-19. [Crossref] [PubMed]
- Wu K, Zhu L, Wang J, et al. A phase II study of concurrent nab-paclitaxel/carboplatin combined with thoracic radiotherapy in locally advanced squamous cell lung cancer. J Thorac Dis 2019;11:4529-37. [Crossref] [PubMed]
- Gandara DR, Hammerman PS, Sos ML, et al. Squamous cell lung cancer: from tumor genomics to cancer therapeutics. Clin Cancer Res 2015;21:2236-43. [Crossref] [PubMed]
- Detterbeck FC, Boffa DJ, Kim AW, et al. The Eighth Edition Lung Cancer Stage Classification. Chest 2017;151:193-203.
- Hwang JK, Page BJ, Flynn D, et al. Validation of the Eighth Edition TNM Lung Cancer Staging System. J Thorac Oncol 2020;15:649-54.
- NCCN Clinical Practice Guidelines in Oncology. Non-Small Cell Lung Cancer Version 3.2020 [database on the Internet]. National Comprehensive Cancer Network (NCCN). 2020. Accessed: March 3, 2020.
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Meng H, Guo X, Sun D, et al. Genomic Profiling of Driver Gene Mutations in Chinese Patients With Non-Small Cell Lung Cancer. Front Genet 2019;10:1008. [Crossref] [PubMed]
- Ding Y, Zhang L, Guo L, et al. Comparative study on the mutational profile of adenocarcinoma and squamous cell carcinoma predominant histologic subtypes in Chinese non-small cell lung cancer patients. Thorac Cancer 2020;11:103-12. [Crossref] [PubMed]
- Pezzella F. Early squamous cell lung carcinoma: prognostic biomarkers for the many. Thorax 2019;74:527-8. [Crossref] [PubMed]
- Shi YX, Wang Y, Li X, et al. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics 2017;18:901. [Crossref] [PubMed]
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 2016;8:a019505. [Crossref] [PubMed]
- Shi YX, Sheng DQ, Cheng L, et al. Current Landscape of Epigenetics in Lung Cancer: Focus on the Mechanism and Application. J Oncol 2019;2019:8107318. [Crossref] [PubMed]
- Dammann R, Strunnikova M, Schagdarsurengin U, et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer 2005;41:1223-36. [Crossref] [PubMed]
- Fukasawa M, Kimura M, Morita S, et al. Microarray analysis of promoter methylation in lung cancers. J Hum Genet 2006;51:368-74. [Crossref] [PubMed]
- Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A 2008;105:252-7. [Crossref] [PubMed]
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics 2009;1:239-59. [Crossref] [PubMed]
- Carvalho RH, Hou J, Haberle V, et al. Genomewide DNA methylation analysis identifies novel methylated genes in non-small-cell lung carcinomas. J Thorac Oncol 2013;8:562-73. [Crossref] [PubMed]
- Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics 2017;12:416-32. [Crossref] [PubMed]
- Pfeifer GP. Defining Driver DNA Methylation Changes in Human Cancer. Int J Mol Sci 2018;19:1166. [Crossref] [PubMed]
- How Kit A, Nielsen HM, Tost J. DNA methylation based biomarkers: practical considerations and applications. Biochimie 2012;94:2314-37. [Crossref] [PubMed]
- Šutić M, Motzek A, Bubanović G, et al. Promoter methylation status of ASC/TMS1/PYCARD is associated with decreased overall survival and TNM status in patients with early stage non-small cell lung cancer (NSCLC). Transl Lung Cancer Res. 2019;8:1000-15. [Crossref] [PubMed]
- Ooki A, Maleki Z, Tsay JJ, et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non-Small Cell Lung Cancer and Serum DNA. Clin Cancer Res 2017;23:7141-52. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Revelo AE, Martin A, Velasquez R, et al. Liquid biopsy for lung cancers: an update on recent developments. Ann Transl Med 2019;7:349. [Crossref] [PubMed]
- Li L, Fu K, Zhou W, et al. Applying circulating tumor DNA methylation in the diagnosis of lung cancer. Precision Clinical Medicine 2019;2:45-56. [Crossref]
- Chae YK, Oh MS. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. J Thorac Oncol 2019;14:16-24. [Crossref] [PubMed]
- Xia S, Ye J, Chen Y, et al. Parallel serial assessment of somatic mutation and methylation profile from circulating tumor DNA predicts treatment response and impending disease progression in osimertinib-treated lung adenocarcinoma patients. Transl Lung Cancer Res 2019;8:1016-28. [Crossref] [PubMed]
- Belinsky SA. Gene Promoter Methylation in Plasma and Sputum Increases with Lung Cancer Risk. Clinical Cancer Research 2005;11:6505-11. [Crossref] [PubMed]
- Wang YC, Hsu HS, Chen TP, et al. Molecular diagnostic markers for lung cancer in sputum and plasma. Ann N Y Acad Sci 2006;1075:179-84. [Crossref] [PubMed]
- Hsu HS, Chen TP, Hung CH, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 2007;110:2019-26. [Crossref] [PubMed]
- Ostrow KL, Hoque MO, Loyo M, et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin Cancer Res 2010;16:3463-72. [Crossref] [PubMed]
- Kneip C, Schmidt B, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol 2011;6:1632-8. [Crossref] [PubMed]
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [Crossref] [PubMed]
- Hulbert A, Jusue-Torres I, Stark A, et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin Cancer Res 2017;23:1998-2005. [Crossref] [PubMed]
- Liang W, Zhao Y, Huang W, et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics 2019;9:2056-70. [Crossref] [PubMed]
- Yang Y, Zheng D, Wu C, et al. Detecting Ultralow Frequency Mutation in Circulating Cell‐Free DNA of Early‐Stage Nonsmall Cell Lung Cancer Patients with Unique Molecular Identifiers. Small Methods 2019;3:1900206. [Crossref]
- Pedersen BS, Eyring K, De S, et al. Fast and accurate alignment of long bisulfite-seq reads. arXiv:14011129v2 [q-bioGN] 2014.
- Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014;30:2503-5. [Crossref] [PubMed]
- Tarasov A, Vilella AJ, Cuppen E, et al. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032-4. [Crossref] [PubMed]


