
Hsa_circ_0006571 promotes spinal metastasis through sponging microRNA-138 to regulate sirtuin 1 expression in lung adenocarcinoma
Introduction
Lung carcinoma is the primary cause of cancer-related mortality worldwide, accounting for almost 20% of all deaths caused by tumors (1,2). More than 65% of patients with lung carcinoma have local or disseminated metastatic disease at diagnosis (3,4). Tumor cells can metastasize locally to the lymph nodes and distant organs, including bone. The spine is the most common site of bone metastasis in lung cancer, with 42% of patients with bone metastasis developing spinal metastasis (SM), followed by the ribs (27%); however, metastasis rarely occurs in appendicular bones (<6%) (5,6). SM is viewed as a deleterious stage which results in a series of skeletal-associated adverse events, especially paraplegia and lifelong urinary and fecal incontinence.
In recent years, studies have increasingly verified that aberrant gene expression due to genomic instability or alteration of epigenetic markers can play a major role in lung cancer metastasis (7,8). As with most malignant tumors, lung cancer metastasis is usually associated with a specific genetic chromosome abnormality (9).
Circular RNAs (circRNAs) and microRNAs (miRNAs) are non-coding RNAs (ncRNAs) that have been studied within cancer context in recent years (10,11). CircRNA are highly stable ncRNAs generated by non-canonical splicing of pre-mRNAs (12). CircRNAs can act as miRNA sponges regulating the function of miRNAs in health and disease (13). For instance, circC3P1 inhibits liver cancer progression through the miR-4641/PCK1 signaling pathway (14). Some researchers have demonstrated that circMAN2B2 regulates the miR-1275/FOXK1 pathway, thereby inducing the progression of lung cancer (15).
MiRNAs are important non-coding RNAs that exert their post-transcriptional regulatory effects by regulating protein-coding gene transcripts (16). Through targeting multiple mRNA transcripts, miRNAs can regulate a variety of biological processes, including migration, apoptosis, and cell proliferation. Notably, the deregulation of miRNA expression through the sponge function of circRNAs may be crucial in cancer etiology (17).
In our study, we discovered lower expression of tumor suppressor miR-138 in SM tissues compared with lung adenocarcinoma, and verified the results using lung cancer cell lines. H1975 and SPA-1 cells migration and invasion was inhibited by the overexpression of miR-138 in vitro. By microarray analysis, we revealed that hsa_circ_0006571, generated from the back-splicing of special AT-rich sequence binding protein 2 (SATB2) gene transcript, was significantly upregulated in SM tissues. Furthermore, we demonstrated that hsa_circ_0006571 could inhibit proliferation and metastatic progression of lung cancer cells via binding to miR-138 as a miRNA sponge to regulate sirtuin 1 (Sirt1) expression. Therefore, hsa_circ_0006571 may have a potential therapeutic effect on lung cancer patients.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1250).
Methods
Patients and tissue samples
Twenty-five cancer lung adenocarcinoma tissues and SM tissues were obtained from the Department of Surgery, Zhongshan Hospital affiliated to Fudan University. Five pairs of samples (lung adenocarcinoma tumor tissues and SM tissues) were used for circRNA microarray, and 20 pairs were used for RT-quantitative polymerase chain reaction (qPCR) validation. All tumor tissues were snap-frozen in liquid nitrogen promptly after surgical excision and were immediately transferred to a −80 °C freezer. The diagnosis of lung adenocarcinoma and SM was pathologically confirmed. None of the patients in our study had received radiotherapy or chemotherapy, or any other cancer therapy before surgery. The study was approved by the Ethics Committee of Zhongshan Hospital, and written informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Expression profile analysis of circRNAs
The microarray “SBC Human ceRNA microarray v1.0” (Biotechnology Corporation, Shanghai, China) was used, which detected 88,371 circRNAs. RNA samples from each group were then used to generate biotinylated cRNA targets for the SBC human ceRNA array V1.0. Biotinylated cRNA targets were then hybridized to the slides. After hybridization, slides were scanned with an Agilent Microarray Scanner (Agilent Technologies, Santa Clara, CA, USA). Microarray experiments were performed by an Agilent Technologies Inc. protocol at Shanghai Biotechnology Corporation.
Cell culture
Human lung adenocarcinoma cell lines, H1975, A549, H1395, and SPC-A1, and a control human bronchial epithelial (HBE) cell line were obtained from the Chinese Academy of Sciences (Shanghai, China). They were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS; Gibco, USA) at 37 °C with 5% CO2.
RNA oligoribonucleotides and cell transfection
Lentiviral vectors for small hairpin RNAs targeting hsa_circ_0006571 were produced by Hanyin, China. H1975 cells were transfected with the corresponding lentiviruses with 8 µg/mL polybrene. Puromycin (4 µg/mL) was introduced to select H1975 cells with stable hsa_circ_0006571 knockdown after 24 h. siRNAs targeting the hsa_circ_0006571 splice junction, Sirt1 mRNA, and miR-138 mimic, miR-138 inhibitor, and their respective controls were synthesized by RiboBio (Guangzhou, China). The siRNA sequences were listed in Table S1.
RNA immunoprecipitation (RIP) and RNA pull-down
Briefly, an RIP assay using SPC-A1 cells was performed with a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). Cells were transfected with Flag-Ago2 lentivirus (Genechem, Shanghai, China) and then treated with Radio Immunoprecipitation Assay (RIPA) Lysis Buffer. The cell lysates were incubated with antibodies against Ago2 (Millipore) or normal mouse immunoglobulin G (Millipore). After being treated with buffer, the immunoprecipitated RNAs were extracted from the cell lysates, and western blotting and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis were then performed.
An RNA pull-down assay was conducted to verify the relationship between hsa_circ_0006571 and miR-138 using the Pierce Magnetic RNA-Protein Pull-Down Kit (BersinBio, Guangzhou, China). For RNA pull-down, specific probes were used to hybridize hsa_circ_0006571, which was pulled down and identified through qRT-PCR. SPC-A1 cells were washed with phosphate-buffered saline (PBS) and lysed with buffer. A biotinylated antisense probe and a control probe were added to the pull-down system before the probes were denatured and hybridized. Non-specifically bound RNA was removed and miRNAs specifically interacting with circRNAs were recovered using Trizol reagent. After reverse transcription, qRT-PCR was performed to testify binding strength.
Cell Counting Kit-8 (CCK-8) assays
Cells were transfected with siRNA, miR-138 mimic, miR-138 inhibitor, and si-hsa_circ_000657 and CCK-8 (Beyotime, China) to test cell viability. After 2 h of incubation, the density values were measured at 450 nm.
Transwell migration assays
Transwell migration assays were performed using Matrigel invasion chambers (8 µm size, Corning, NY, USA) following the manufacturer’s instructions. A total of 3×104 H1975 and SPA-1 cells were resuspended in 200 µL serum-free DMEM, and then seeded into the upper chambers of Transwell plates, respectively. Complete DMEM medium with 5% FBS was placed into the lower chamber. Cells adhering to the membrane were fixed in 4% paraformaldehyde for 15 minutes, stained with crystal violet, and counted under a microscope.
Cell cycle assays
After transfection and culture, A total of 3×105 H1975 and SPCA-1 cells were harvested and suspended in 500 µL of binding buffer, respectively. Then, 5 µL of Annexin V-Fluorescein Isothiocyanate/propidium iodide (V-FITC/PI) (Beyotime, Shanghai, China) was added to the cell suspension, and the cells were incubated at 37 °C for 20 min in the dark. Flow cytometry analysis (BD-FACS Calibur, Franklin Lakes, NJ, USA) was performed to detect cell apoptosis and the cell cycle.
Cell apoptosis assay
A total of 105 A549 and SPC-A1 cells were washed with PBS and fixed in 4% paraformaldehyde for 20 minutes. The cells were then incubated with 5 µL Annexin V-phycoerythrin conjugate and 5 µL propidium iodide (Beyotiome, Shanghai, China) for 20 minutes at 37 °C in the dark. The results were analyzed using FlowJo software (version 10.0) (FlowJo, Ashland, OR, USA).
qRT-PCR analysis
The reverse transcription and quantification of miR-138 were performed with a miRNA qRT-PCR Kit (Tiangen Biotech, Beijing, China). RNA was reverse transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Japan). An ABI Prism 7500 RealTime PCR system (Applied Biosystems, Foster City, USA) was used to carry out qPCR using SYBR Premix Ex Taq II (TaKaRa). The 2−ΔΔCt method was used to determine the relative gene expression level. The primers are shown in Table S2.
Luciferase reporter assays
The wild-type (WT) sequences with putative hsa-mir-138 binding site in the three prime untranslated region (3'-UTR) of Sirt1 and its mutant dual-luciferase reporter vectors were constructed by Hanyin, China. Briefly, the 3'-UTR of the human Sirt1 gene was amplified through PCR using the following primer sequence: forward primer: 5'-AACGAGCTCGCTAGCCTCGAGTGTAATAATTGTGCAGGTACAGGAATTG-3'; reverse primer: 5'-ATGTCTGCTCGAAGCGGCCGCGTTAGCTGCCACAGTTTTGGAAAATGC-3'. The PCR product was cloned into the multiple cloning site of the basic dual-luciferase reporter vector pmiGLO, which is located downstream of the luciferase reporter gene, between the XhoI and NotI sites, to generate pmirGLO-Sirt1-WT. The mutant vector was amplified using the following primers: forward primer-1: 5'-TTGTGCAGGTACAGGAATTGTTCTCATGTTATTAGGAACTTTAGCATGTC-3'; reverse primer-1: 5'-ATGTCTGCTCGAAGCGGCCGCGTTAGCTGCCACAGTTTTGGAAAATGC-3'; forward primer-2: 5'-AACGAGCTCGCTAGCCTCGAGTGTAATAATTGTGCAGGTACAGGAATTG-3'; reverse primer-2: 5'-ATGTCTGCTCGAAGCGGCCGCGTTAGCTGCCACAGTTTTGGAAAATGC-3'. The PCR product was inserted into the pmirGLO basic vector.
RNA fluorescence in situ hybridization
The RNA fluorescence in situ hybridization (FISH) assay was performed with a Fluorescent In Situ Hybridization Kit (RiboBio, China). Cy3-labeled hsa_circ_0006571 probes were used to measure with the kit, and then visualized with confocal microscopy.
Cell morphology
H1975 and SPC-A1 cells were planted in a 12-well palate and then transfected with si-hsa_circ_000657. The cells were cultured for 48 h to allow enough time for morphology changes. After 48 h, the plates were washed with PBS to remove dead cells. Finally, H1975 and SPC-A1 cells were observed using an optical microscope (Olympus Corporation, Tokyo, Japan), and changes in cell morphology were analyzed. When H1975 and SPC-A1 cells show less fibroblast-like morphology and cellular protrusions, it indicates that epithelial-mesenchymal transition (EMT) metastasis is inhibited.
Western blotting
A quantitative protein from tissues and cells (20 µg) was injected into Bis-Tris sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) gel and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking for 1 h at 37 °C, the membranes were incubated overnight at 4 °C with the following primary antibodies (CST, Danvers, MA, USA): Sirt1 (1:1,000), Vimentin (1:1,000), E-cadherin (1:1,000), zonula occludens-1 (ZO-1) (1:1,000), Snail (1:1,000), zinc finger E-box binding homeobox 1 (ZEB1) (1:1,000); cleaved caspase-3 (1:1,000) (Beyotime, Shanghai, China); and β-actin antibody (1:1,000) (Beyotime, Shanghai, China). The membranes were then exposed to a horseradish peroxidase (HRP)-labeled secondary antibody, goat anti-rabbit (1:5,000) or goat anti-mouse (1:5,000), for 2 h. The color reaction was observed with Quantity One 4.6.2 software (Bio Rad, Hercules, CA, USA).
Immunohistochemistry
The sections were deparaffinized, rehydrated, and treated with antigen-retrieval buffer and diluted hydrogen peroxide. The slides were incubated overnight at 4 °C with antibodies to Sirt1 and Vimentin (CST, Danvers, MA, USA). Then, the slides were stained with secondary antibodies for 60 min. The positivity of immunoreactivity was evaluated based on the proportion of positive cells.
Xenograft mouse model of SM
Five-week-old nude mice (half males and half females) were randomly divided into two groups and anesthetized using sodium pentobarbital (35–40 mg/kg). Approximately 1×106 sh-circ0006571 and sh-NC H1975 cells were suspended in 200 µL of serum-free medium. Then, cells were injected into the left ventricles of the mice to establish SM animal models as previously described (18). After 4–8 weeks, the mice underwent luciferase signal scanning (Quantum GX, PerkinElmer, Waltham, MA, USA). Mice suspected with SM were subjected to further micro-computed tomography (CT) scanning (Quantum GX, PerkinElmer, USA) and pathological examination. Experiments were performed under a project license (No. 2017-154) granted by Animal Ethics Committee of Zhongshan Hospital, in compliance with national guidelines for the care and use of animals.
Statistical analyses
Data are expressed as the mean ± standard deviation (mean ± SD). Differences between two groups were assessed by performing Student’s t-test. Differences between more than two groups were analyzed with one-way analysis of variance. Fisher’s exact test was used to analyze the results of the in vivo experiments. Statistical analyses were conducted with SPSS software (version 21.0), (SPSS, UK). P<0.05 was considered to show statistical significance.
Results
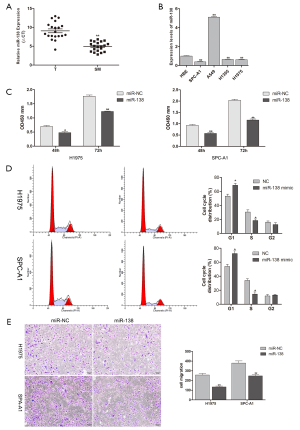
miR-138 inhibited proliferation, migration, and cell-cycle in SPC-A1 and H1975 cells in vitro
The quantification of miR-318 by qRT-PCR in human samples showed that miR-138 was significantly downregulated in SM tissues compared to lung cancer tissues (Figure 1A). Then, the expression of miR-138 in lung adenocarcinoma cell lines (H1975, A549, H1395, and SPC-A1) and an HBE cell line was investigated. The data showed a low expression of miR-138 in H1975 and SPC-A1 cells (Figure 1B). However, the expression level of miR-138 was slightly increased in A549 cell line. This may be due to different cancer cell lines having different genetic backgrounds. To study the function of miR-138 in lung adenocarcinoma cell lines, miR-138 was overexpressed in H1975 and SPC-A1 cells. The proliferation ability of H1975 and SPC-A1 cells was found to be significantly restrained at 48 and 72 h after transfection with miR-138 mimic (Figure 1C). Cell cycle analysis indicated that miR-138 overexpression markedly arrested H1975 and SPC-A1 cells at the G1/S phase (Figure 1D). Also, the migration ability of H1975 and SPC-A1 cells transfected with miR-138 mimic was lower than that of the control cells (Figure 1E). These results show that miR-138 inhibited the proliferation and migration of lung adenocarcinoma cancer cells in vitro.
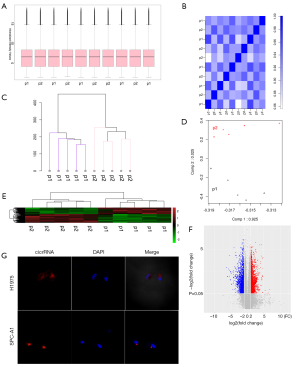
CircRNA expression profile analysis
To identify specific circRNAs that might be involved in SM, five pairs of lung adenocarcinoma tissues and SM tissues pooled from patients were selected. First, the correlation of matched tissues was confirmed using box plots, correlation plots, Hclust plots, and principal component analysis (Figure 2A,B,C,D). Then, the lung adenocarcinoma and SM tissue specimens in five pairs were subjected to circRNA microarray assay analysis after experimental validation. A total of 3,810 circRNAs were found to be differentially expressed with a more than two-fold change between lung adenocarcinoma tissues and SM tissues (Figure 2E,F). Subsequently, FISH assays showed that hsa_circ_0006571 is predominately localized in the cytoplasm (Figure 2G).
miR-138 targeted Sirt1 in lung adenocarcinoma cancer in vitro
Consistent with the previous experimental evidence, we also identified a negative feedback loop between miR-318 and Sirt1 (Figure 3A,B,C). After Sirt1 had been established as a target of miR-138, western blotting was carried out to analyze the expression of proteins involved in tumor cell EMT. The results showed that the expression levels of Sirt1 and Vimentin in H1975 and SPC-A1 cells were clearly decreased after transfection with miR-138 mimic, thus suggesting that miR-138 is involved in the EMT process in tumor cells. Moreover, we performed RIP assay for AGO2 in tumor cells and investigated the expression level of hsa_circ_0006571 and miR-138 pulled-down from AGO2-expressed cells by qRT-PCR analysis. The results indicated that association between hsa_circ_0006571 and miR-138 (Figure 3D). Meanwhile, an RNA pull-down assay detected that hsa_circ_0006571 directly interacted with miR-138. The results demonstrated that compared with control group, miR-138 were significantly enriched in the hsa_circ_0006571 probe group (Figure 3E). Also, the expression of Sirt1 was detected in 10 pairs of samples by qRT-PCR and western blotting. Sirt1 was found to be upregulated in SM tissues compared with lung adenocarcinoma tissues (Figure 3F,G). The results of IHC also validated that Sirt1 and Vimentin were upregulated in SM tissues (Figure 3H).
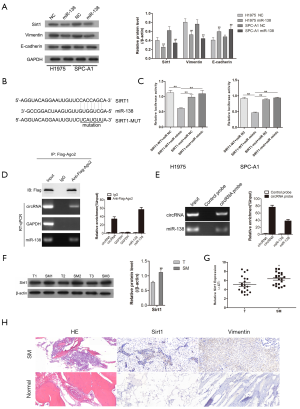
Hsa_circ_0006571 directly targeted miR-138 in cells
To further testify the association between hsa_circ_0006571 and miR-138, RIP assays were performed using an RIP assay Kit (Millipore, Billerica, Massachusetts). Subsequently, SPC-A1 cell extracts were incubated with Ago2 (Millipore) or IgG (Millipore) according to the specifications. The results showed significant enrichment of miR-138 and hsa_circ_0006571 with AGO2 antibody (Figure 3D). Subsequently, the RNA pull-down assay confirmed the interaction between hsa_circ_0006571 and miR-138. The results indicated a higher level of miR-138 in the hsa_circ_0006571 pulled-down pellet than in the negative control (Figure 3E). Both the RIP and RNA pull-down assays confirmed the interaction between hsa_circ_0006571 and miR-138 via Ago2-containing complexes.
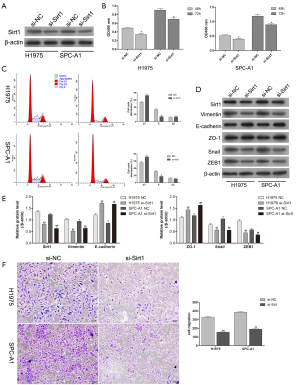
Downregulation of Sirt1 inhibited the proliferation, invasion, and migration abilities of cancer cells
H1975 and SPC-A1 cells were transfected with an si-Sirt1 RNA interference sequence to inhibit Sirt1 expression. At the molecular level, western blotting showed that Sirt1 expression was clearly decreased by the interference sequence, and the downregulation of Sirt1 inhibited H1975 and SPC-A1 cell proliferation (Figure 4A,B). The cell cycle data showed that Sirt1 knockdown arrested H1975 and SPC-A1 at the G1/S phase (Figure 4C). With the downregulation of Sirt1 protein expression, the expression levels of the EMT markers Vimentin, Snail, and ZEB1 decreased, whereas those of E-cadherin and ZO-1 increased (Figure 4D,E). Moreover, the downregulation of Sirt1 decreased the migration ability of H1975 and SPC-A1 cells (Figure 4F). In conclusion, these findings suggested that Sirt1 exerts an oncogenic effect in lung cancer by regulating the EMT process.
hsa_circ_0006571 regulates the proliferation and migration of lung adenocarcinoma cells via the miR-138-Sirt1 axis
To explore the effect of hsa_circ_0006571 on SM, qRT-PCR was performed on 10 pairs of SM tissues to verify the circRNA array data. As miR-138 was downregulated in SM tissues, we speculated that miR-138 may be suppressed in SM by the upregulation of hsa_circ_0006571. Consistent with that of the miR-138 target Sirt1, the expression of hsa_circ_0006571 was found to be upregulated in SM tissues compared with lung adenocarcinoma tissues (Figure 5A).
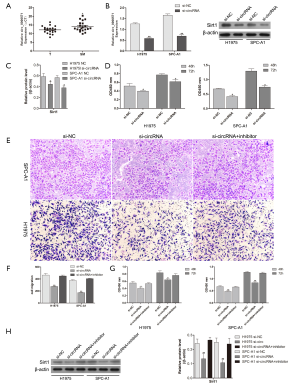
After confirming that hsa_circ_0006571 binds to miR-138 and is positively correlated with Sirt1 expression, we considered that hsa_circ_0006571 may have an oncogenic role in lung adenocarcinoma cells. Hsa_circ_0006571 was knocked down via a specific siRNA targeting the splice junction of hsa_circ_0006571 (Figure 5B,C). The proliferation ability of H1975 and SPC-A1 cells was significantly decreased by si-hsa_circ_0006571 (Figure 5D).
However, no apparent increase in miR-138 levels was observed as a result of hsa_circ_0006571 knockdown. Rescue experiments were conducted to identify whether hsa_circ_0006571 regulates H1975 and SPC-A1 cell proliferation and Sirt1 expression through inhibiting miR-138. The decreases in the proliferation and migration of H1975 and SPC-A1 cells resulting from hsa_circ_0006571 knockdown was rescued by the addition of miR-138 inhibitor (Figure 5E,F,G). Moreover, the repression of Sirt1 by hsa_circ_0006571 was also rescued after miR-138 was inhibited (Figure 5H). These results suggest that hsa_circ_0006571 regulates the proliferation and migration of lung adenocarcinoma cells via the miR-138-Sirt1 axis.
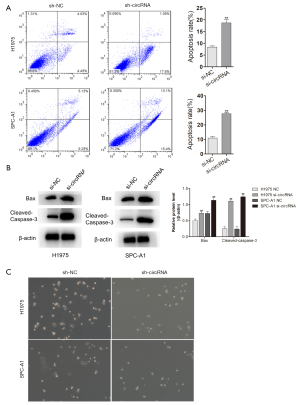
hsa_circ_0006571 inhibited cell apoptosis and promotes EMT in H1975 and SPCA-1 cells
To further test the effects of hsa_circ_0006571 on H1975 and SPCA-1 cells, siRNAs were used to downregulate the expression of hsa_circ_0006571. Flow cytometry was performed to detect cell apoptosis, and downregulation of hsa_circ_0006571 was found to promote cell apoptosis, which was consistent with the results of western blot (Figure 6A,B). Further, cells treated with siRNA exhibited morphological changes. Cells lost their elongated fibroblast-like shape and acquired an epithelial cobblestone-like morphology. After transfection, cellular protrusions disappeared when compared to control group (Figure 6C). These modifications were observed in both H1975 and SPC-A1 cells and indicated that the downregulation of hsa_circ_0006571 inhibited the EMT process.
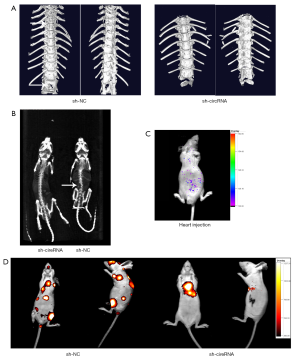
Knockdown of hsa_circ_0006571 suppressed tumor growth in vivo
In vitro, hsa_circ_0006571 knockdown was observed to suppress the viability of H1975 and SPC-A1 cells. Therefore, we postulated that hsa_circ_0006571 may play the same role in vivo. H1975 cells with stable hsa_circ_0006571 knockdown (sh-hsa_circ_0006571-H1975) or cells infected with control vector (sh-NC-H1975) were injected into the left ventricle of nude mice.
Consistent with the in vitro results, knockdown of hsa_circ_0006571 suppressed SM in vivo. Micro CT, Spect-CT, and images of luciferase signal scans showed SM in 4 of the 10 nude mice in the si-H1975 group, and the lesion of each mouse involved multiple vertebral segments. In the sh-NC-H1975 group, 1 of the 10 nude mice exhibited SM (Figure 7 and Figure 8A). IHC analysis also validated the downregulation of Sirt1 and Vimentin in sh-hsa_circ_0006571-H1975 samples in vivo (Figure 8B).
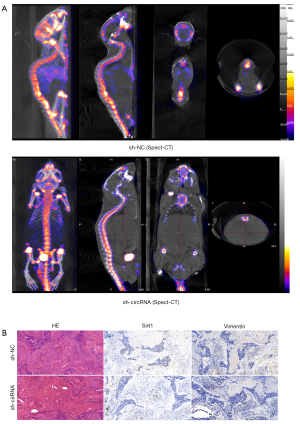
The differences in the occurrence of SM between the sh-hsa_circ_0006571-H1975 and sh-NC-H1975 groups were not statistically significant. All SM samples were examined through pathological examination, including hematoxylin-eosin staining and IHC. In conclusion, hsa_circ_0006571 may act as a novel therapeutic target for SM of lung adenocarcinoma tumors via the miR-138-Sirt1 axis.
Discussion
MiRNAs are important epigenetic regulators of gene expression which are involved in cancer metastasis. MiRNAs participate in multi-level processes and play a key role in the progression of tumors (19). Here, we confirmed that miR-138 was downregulated in SM samples and inhibited the proliferation and migration abilities of SPC-A1 and H1975 cells. This is in agreement with the findings of previous research that demonstrated the anti-oncogenic function of miR-138 in multiple cancers (20).
Generally, miRNAs bind the 3'-UTRs of messenger RNAs (mRNAs), leading to the degeneration of mRNAs or translational blockage (21). The current study validated Sirt1 as a target of miR-138 and verified its role in lung adenocarcinoma. Sirt1 has important involvement in cell proliferation and apoptosis, especially in tumorigenesis and cancer metastasis (22,23). Sirt1 could interact with p53 and result in deacetylation of Lys 382 at the C-terminal of p53. Therefore, a series of tumor suppressor were not activated by p53, such as transcription activation (24). In epigenetics, Sirt1 also could inhibit the tumor suppressor functions and affect the activities of DNA repair-related protein (25,26). Our in vitro experiments demonstrated that miR-138 overexpression downregulated Sirt1 expression by facilitating the degradation of Sirt1. In agreement with previous findings, our functional experiments revealed that silencing of Sirt1 decreased cell proliferation, migration and inhibited the EMT process. This resulted in the upregulation of E-cadherin and ZO-1, and the downregulation of Vimentin, snail, and ZEB1, which are involved in EMT (27-29).
CircRNAs are increasingly being reported to be important regulators in various physiological and pathological cancer processes. Dysregulation of circRNA expression contributes to malignant behaviors in cancers, including lung cancer (30), hepatocellular cancer (31), and breast cancer (32). In summary, dysregulated circRNA expression is clearly associated with the progression and prognosis of cancer, and is always directly involved in malignant behaviors, including proliferation, metastasis, and invasion (33). Uncovering the relationship between circRNA and spine metastasis of lung cancer is crucial. Considering the downregulation of miR-138 in SM tissues,and the and role of circRNAs, we hypothesized that the low expression level of miR-138 may partly be attributed to the abundance of circRNAs. In this study, we used the SBC human ceRNA array V1.0 to identify a circRNA termed hsa_circ_0006571 and validate its tumor oncogenic function in lung cancer metastasis. H1975 and SPC-A1 cells transfected with si-hsa-circ_000657 were observed to display evident morphological changes, from an elongated morphology with cellular protrusions to an epithelial cobblestone-like morphology, which indicated the inhibition of the EMT process. This suggested that si-hsa-circ_000657 could inhibit cells to gain a fibroblast-like mesenchymal morphology, one of the main features acquired by cells undergoing EMT, and could then reduce spinal metastatic potential of lung cancers. Therefore, hsa_circ_0006571 may serve as a potential biomarker for lung adenocarcinoma diagnosis and prognosis.
CircRNAs often act as miRNA sponges. To explore the functional mechanism of circRNAs and miRNA-138, we used circRNA microarray to determine potential circRNAs, and hsa_circ_0006571 was eventually identified. Through luciferase reporter assays and RNA FISH assays, the association between hsa_circ_0006571 and miR-138 in lung adenocarcinoma cells was identified. Meanwhile, RIP and RNA pull-down showed that hsa_circ_0006571 served as a miRNA sponge for miR-138 in H1975 and SPC-A1 cells. It is worth noting that the high expression level of hsa_circ_0006571 was positively correlated with the high expression of Sirt1, which indicated that hsa_circ_0006571 might regulate Sirt1 expression by sponging miR-138. The consequent declines in the protein level of Sirt1, and in cell viability and migration were rescued by inhibiting miR-138, which was consistent with the findings of previous studies. This result indicated that hsa_circ_0006571 regulates Sirt1 expression via miR-138. However, in our in vivo experiments, no statistically significant differences in SM were observed between the sh-hsa_circ_0006571-H1975 and sh-NC-H1975 groups. This lack of significance might have resulted from the limited sample size or because inoculated cells did not undergo bone tropism induction, as described in a previous study (18); however, the rates of SM and the involvement of pathological vertebral segments were increased in the sh-NC-H1975 group compared to the sh-hsa_circ_0006571-H1975 group. Additionally, we detected the correlations among hsa_circ_0006571, miR-138, and Sirt1 in cancer tissues, and the underlying mechanism, by using techniques including western blotting and luciferase assay. In future studies, we may investigate the mechanism of SM of lung cancer using cancer cells overexpressing hsa_circ_0006571 combined with bone tropism induction. Overall, our experiments revealed that hsa_circ_0006571 regulates the proliferation and migration of lung adenocarcinoma cells via the miR-138-Sirt1 axis.
In conclusion, this study provides initial evidence that hsa_circ_0006571 regulates the migration and proliferation of lung adenocarcinoma cells via the miR-138-Sirt1 axis. Our in vivo experiment also showed that hsa_circ_0006571 knockdown can suppress the development of SM. This study sheds light on a novel mechanism potentially involved in SM of lung adenocarcinoma.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: This work was supported by the National Natural Science Foundation of China (81572629, 81772855 and 81701370).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1250
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1250
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1250). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Zhongshan Hospital, and written informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. 2017-154) granted by Animal Ethics Committee of Zhongshan Hospital, in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Devesa SS, Bray F, Vizcaino AP, et al. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer 2005;117:294-9. [Crossref] [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Zheng XQ, Huang JF, Lin JL, et al. Incidence, prognostic factors, and a nomogram of lung cancer with bone metastasis at initial diagnosis: a population-based study. Transl Lung Cancer Res 2019;8:367-79. [Crossref] [PubMed]
- Rami-Porta R, Crowley JJ, Goldstraw P. The revised TNM staging system for lung cancer. Ann Thorac Cardiovasc Surg 2009;15:4-9. [PubMed]
- Sugiura H, Yamada K, Sugiura T, et al. Predictors of survival in patients with bone metastasis of lung cancer. Clin Orthop Relat Res 2008;466:729-36. [Crossref] [PubMed]
- Tsuya A, Kurata T, Tamura K, et al. Skeletal metastases in non-small cell lung cancer: a retrospective study. Lung Cancer 2007;57:229-32. [Crossref] [PubMed]
- Goto T, Hirotsu Y, Mochizuki H, et al. Mutational analysis of multiple lung cancers: Discrimination between primary and metastatic lung cancers by genomic profile. Oncotarget 2017;8:31133-43. [Crossref] [PubMed]
- Illei PB, Belchis D, Tseng LH, et al. Clinical mutational profiling of 1006 lung cancers by next generation sequencing. Oncotarget 2017;8:96684-96. [Crossref] [PubMed]
- Guan ZB, Cao YS, Li Y, et al. Knockdown of lncRNA GHET1 suppresses cell proliferation, invasion and LATS1/YAP pathway in non small cell lung cancer. Cancer Biomark 2018;21:557-63. [Crossref] [PubMed]
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016;1859:163-8. [Crossref] [PubMed]
- Yang X, Su W, Chen X, et al. Validation of a serum 4-microRNA signature for the detection of lung cancer. Transl Lung Cancer Res 2019;8:636-48. [Crossref] [PubMed]
- Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 2014;15:9331-42. [Crossref] [PubMed]
- Li RC, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis 2018;9:838. [Crossref] [PubMed]
- Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun 2018;499:1044-9. [Crossref] [PubMed]
- Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun 2018;498:1009-15. [Crossref] [PubMed]
- Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as Biomarkers and Therapeutic Targets in Non-Small Cell Lung Cancer: Current Perspectives. Target Oncol 2017;12:179-200. [Crossref] [PubMed]
- Jin X, Feng CY, Xiang Z, et al. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 2016;7:66455-67. [Crossref] [PubMed]
- Zadnik P, Sarabia-Estrada R, Groves ML, et al. A novel animal model of human breast cancer metastasis to the spine: a pilot study using intracardiac injection and luciferase-expressing cells. J Neurosurg Spine 2013;18:217-25. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Manafi Shabestari R, Alikarami F, Bashash D, et al. Overexpression of MiR-138 Inhibits Cell Growth and Induces Caspase-mediated Apoptosis in Acute Promyelocytic Leukemia Cell Line. Int J Mol Cell Med 2018;7:24-31. [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- Liu L, Liu C, Zhang Q, et al. SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology 2016;64:814-27. [Crossref] [PubMed]
- Shen ZL, Wang B, Jiang KW, et al. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget 2016;7:35092-105. [Crossref] [PubMed]
- Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001;107:149-59. [Crossref] [PubMed]
- Dai JM, Wang ZY, Sun DC, et al. SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J Cell Physiol 2007;210:161-6. [Crossref] [PubMed]
- Wang C, Chen L, Hou X, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol 2006;8:1025-31. [Crossref] [PubMed]
- Guo SJ, Zeng HX, Huang P, et al. MiR-508-3p inhibits cell invasion and epithelial-mesenchymal transition by targeting ZEB1 in triple-negative breast cancer. Eur Rev Med Pharmacol Sci 2018;22:6379-85. [PubMed]
- Liu Z, Chen J, Yuan W, et al. Nuclear factor I/B promotes colorectal cancer cell proliferation, epithelial-mesenchymal transition and 5-fluorouracil resistance. Cancer Sci 2019;110:86-98. [Crossref] [PubMed]
- Zhao JL, Liang SQ, Fu W, et al. The LIM domain protein FHL1C interacts with tight junction protein ZO-1 contributing to the epithelial-mesenchymal transition (EMT) of a breast adenocarcinoma cell line. Gene 2014;542:182-9. [Crossref] [PubMed]
- Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer 2018;17:140. [Crossref] [PubMed]
- Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle 2018;17:2349-59. [Crossref] [PubMed]
- Zhang HD, Jiang LH, Hou JC, et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics 2018;10:1229-42. [Crossref] [PubMed]
- Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol 2017;10:2. [Crossref] [PubMed]


