
Narrative review: molecular and genetic profiling of oligometastatic non-small cell lung cancer
Introduction
Rationale
Traditionally, using established methods of cancer staging, lung cancer has been broadly categorized into localized and metastatic disease (1). According to expert guidelines for advanced non-small cell lung cancer (NSCLC), whether the burden of metastatic disease is limited or widespread, these cases fall into the same category of incurable disease (2). More recently, there has been growing recognition of a subset of patients who have limited number of metastases (“oligometastatic disease”), who demonstrate long-term survival and, in some cases, curative outcomes when the primary tumor and the metastatic sites are treated with localized therapy (3-5). Interest in an aggressive local therapy approach for oligometastatic NSCLC has grown over the last decade. Factors leading to this interest include: improved imaging technology to better characterize sites of disease leading to upstaging of some cases [positron emission tomography (PET), computed tomography (CT), and magnetic resonance imaging (MRI)]; the introduction of better systemic therapies to control micro-metastatic disease; and better local therapy options including minimally invasive surgery and stereotactic radiation therapy.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-448).
Objectives
In this review we will discuss: the definition, clinical and biologic features of oligometastatic NSCLC, as well as the concept of treating oligoprogression in oligometastatic NSCLC.
Methods
After a systematic review of the literature, a PubMed search was performed using the English language and the key terms: Oligometastatic, non-small cell lung cancer (NSCLC), localized consolidative treatment (LCT), biomarkers, biologic features, clinical features. Over 500 articles were retrieved between 1889–2021. A total of 178 papers (ranging from 1907 until 2021) discussing the definition, clinical and biologic factors leading to oligometastatic cancer with a focus on NSCLC were reviewed and included for discussion of this paper.
The mechanism of metastatic progression
It is now clear that the metastatic process is more complex than the orderly progression from primary tumor to regional lymph nodes and then distant organs initially proposed in the 1800s (6). According to the seed and soil hypothesis formulated in the late 1980s (7), metastases are not only driven by the circulatory patterns but rather a predetermined process that depends on the microenvironments of both the primary tumor and the target organ. In 1995 Helman and Weichselbaum postulated that metastases develop over several consecutive steps correlating with the biology of tumor progression, from localized disease that is curable with localized ablative approach, to limited metastatic disease in limited organs which is amenable to improved survival by localized therapy, to diffuse metastases (8).
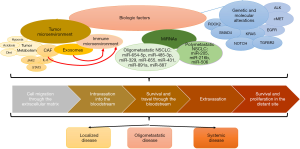
Cancer cells that become metastatic have characteristics that distinguish them from other cancer cells in the primary tumor. To colonize new organs, these cells experience loss of cellular adhesion, intravasation, survival in the circulation, and extravasation (9,10). Cancer cells that remain localized usually lack one or more of these features (Figure 1) (11). Primary tumors may contain certain subsets of tumor cells or clones that are predisposed to metastasize. These subsets/clones may not necessarily aid in the growth of the primary tumor but can promote the development of metastases at a very early stage through hematogenous dissemination (12,13). This suggests that there are some patients who have a disease that is clinically metastatic, but would result in only a few metastases (biologically is oligometastatic) and might benefit from aggressive local therapy and of course tumors that are prone to have wide spread metastases. The potential characteristics of these two are summarized in Table 1 (14).

Full table
The definition of oligometastatic disease
Clinically, oligometastatic disease is defined as a subtype of metastatic disease that is limited in total disease burden by the number of metastatic sites (15). Biologically, oligometastatic disease may represent a distinct category with specific molecular characteristics which results in a disease state in between localized and widespread systemic disease (Table 1). Currently, there is no consistent clinical distinction between oligometastatic disease and diffuse metastatic disease. While some studies define limited number of metastases as being six or fewer metastatic sites, other studies define oligometastatic disease as ≤5 metastases, while still others use a threshold of ≤4 sites (16). This definition will likely continue to mature through clinical trials and biomarker studies and the effect of localized consolidative approaches will be better characterized. For NSCLC, this represents an important area of investigation, as it is estimated that up to 50% of patients with metastatic have a limited number of metastases (17).
Clinical features of oligometastatic disease
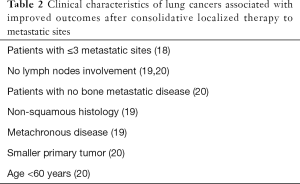
Using contemporary, highly sensitive imaging techniques, we can now reliably characterize cancers according to tumor size, number of metastases, and disease-free interval. Among cases that meet one common clinical criterion of oligometastatic disease (i.e., ≤6 sites), larger primary tumors, greater number of visualized metastases, more advanced mediastinal lymph node involvement, synchronous metastases, advanced age, bone metastatic disease and squamous histology are associated with higher likelihood of occult, micro-metastatic disease and thus higher risk of recurrence after localized consolidative therapy (Table 2) (18-22). Conversely, patients who have longer disease-free interval and slower pace of dissemination typically have lower risk of recurrence after localized consolidative therapy (23,24).
Full table
While these clinical features can help in the selection process for patients who would benefit from localized therapy to metastatic sites, in some cases they may inaccurately categorize a systemic disease with limited number of visible metastases and diffuse occult metastases as oligometastatic disease. Such patients typically have poor outcomes despite aggressive localized treatment to clinically apparent oligometastatic sites. On the other hand, some patients considered high risk for recurrence based on clinical features may derive long-benefit from localized treatments. Accordingly, there is growing interest in biologic factors underlying these distinct and unpredictable scenarios.
Biologic factors associated with oligometastatic disease
Although localized therapy for oligometastatic disease with surgery and/or radiation has been offered to patients for decades (3-5), clinical trials establishing this treatment strategy generally have not explored the molecular characteristics of these cases (25). Currently, the selection of oligometastatic patients for such trials is primarily based on the number of metastases present, and the length of the disease-free interval (26). Improved understanding of the molecular features, expression signatures and other biologic hallmarks might allow a more reliable selection of patients who could benefit from oligometastatic guided therapy (27). Although studies of biologic factors in NSCLC are limited, more extensive data from breast cancer, prostate cancer, ovarian cancer, and other malignancies may provide relevant insights. Such studies have suggested that numerous factors are associated with tumor metastatic potential, including genomic alterations, epigenetic modifications, tumor metabolism, tumor microenvironment, microRNAs (miRNAs), and immune signature (Figure 1). In the following sections, we review each of these categories.
Genomic alterations
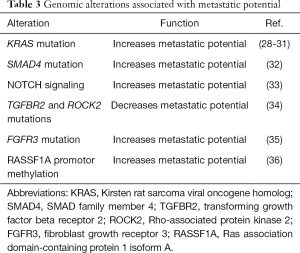
In lung cancer, certain genomic alterations are linked to more aggressive tumors, higher metastatic potential, and worse survival, while other signatures are linked to slower growth and oligometastatic disease (Table 3).
Full table
To elucidate the cellular dynamics in lung adenocarcinoma progression, comprehensive single-cell transcriptome profiling of primary and metastatic samples has been performed (37). A pattern consistent with aggressive cell movement and abnormal proliferation or apoptosis was predominantly identified in cells isolated from the late-stage biopsies or metastases, suggesting an association with tumor progression and metastasis. Furthermore, patients with this signature had worse overall survival.
Tumor microenvironment
Tumor metabolism
The metabolic microenvironment can influence tumor stroma and affect the metastatic potential. It has been suggested that changes in the metabolism surrounding the primary cancer cells can dictate its metastatic potential (38). For example, a high sugar diet, obesity, and a diet high in fat has been shown to promote metastatic potential in several types of cancer (39-42). Contrary to normal cells most cancer cells depend on aerobic glycolysis instead of mitochondrial oxidative phosphorylation as the energy source, and thus cancer cells have increased glucose uptake and glycolysis utilization leading to lactate production which is also known as the “Warburg effect” (43). Fructose derived from the sucrose was found to be responsible for the development of lung metastases through induction of 12-LOX signaling (39). Conversely, a decrease in fructose consumption limits metastatic potential (44). Obesity has also shown to promote metastasis through increased lipogenesis, increased vascularity and decreased M1/M2 macrophage ratios which accounts for enhanced tumorigenicity (41).
Under hypoxia and acidosis, cancer cells exhibit increased metastatic potential and that is mediated by proteoglycan-dependent endocytosis (45). In diabetic patients, hyperglycemia can impair tumor growth in early stages via attenuation of angiogenesis; however, this biology may also enhance metastatic seeding through neutrophil impairment with reduced production of granulocyte-colony stimulating factor (G-CSF) (46).
Cancer-associated fibroblasts (CAFs)
In NSCLC, CAFs can modulate cancer cell proliferation, angiogenesis, invasion, and metastasis through interplay with tumor cells and the tumor microenvironment (47). CAFs isolated from human lung cancer tissues secrete interleukin-6 (IL-6), which stimulates Janus kinase 2 (JAK2)-signal transducer and activates signal transducer and activator of transcription 3 (STAT3) signaling in human lung cancer cells to increase metastasis in vivo (48). CAFs also drive expression of α11β1 integrin, leading to increased extracellular matrix stiffness that contributes to increased growth and metastasis of patient-derived xenografts (49). On the other hand, CAFs can modulate immune responses in the tumor microenvironment and are able to cross-present antigens complexed with major histocompatibility complex class I (MHC I) to antigen-specific CD8+ T cells (50). CAFs may also enhance the recruitment and activation of anti-tumor CD8+ T cells through expression of CCL19. This interaction between fibroblasts and immune cells can strongly restrict cancer progression through an enhanced immune response (51). The detection of CAFs in the blood is linked to metastatic disease in breast cancer and can be investigated as promising biomarker for metastasis in lung cancer (52).
MiRNAs
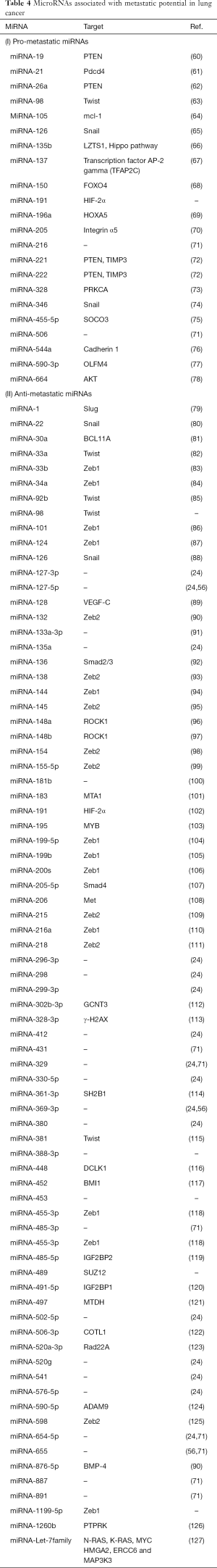
MiRNAs are a class of small, noncoding RNAs that suppress gene expression through direct interaction with the target messenger RNA (53). They have been demonstrated to regulate multiple steps in the metastatic cascade, including epithelial-mesenchymal transition (EMT), a process that promotes motility and invasiveness, and allows cancer cells to detach from the primary tumor and relocate to a secondary site (54-56). MiRNA expression profiling of tumor samples may accurately distinguish between patients with limited metastatic disease who are truly oligometastatic versus those who will likely develop widespread metastatic disease later (24,57). Lussier and colleagues analyzed the miRNA patterns in samples taken from resected lung cancer metastases from patients with oligometastatic cancers and found that a specific set of miRNAs that are known to be associated with tumor-suppression functions were down-regulated in a group of patients with a high rate of progression (24). They also investigated miRNA profiles and expression patterns in primary and metastatic samples from cancer patients and found that high expression of miR-200c in metastatic tumors resulted in a significant increase in the metastatic burden and was shown to predict progression towards polymetastases through regulation of EMT-related pathways (57).
Patients with NSCLC with and without metastasis, exhibit different miRNA profiles (58,59). Specific miRNAs have been found to control certain functional pathways and thus believed to contribute to the lung cancer metastatic potential: hsa-let-7a (inhibits cell proliferation through suppression of RAS and repression of the HMGA2 oncogene and associated with prolonged survival in NSCLC), hsa-miR-221 (inhibits angiogenesis in lung cancer), hsa-miR-137 (promotes lung cancer invasion), hsa-miR-372 (promotes tumor proliferation), and hsa-miR-182 (promotes lung cancer invasion) (58) (Table 4). Wang et al. identified a panel of 10 miRNAs that could distinguish the oligo- from polymetastatic lung cancer (71). MiR-654-5p, miR-485-3p, miR-329, miR-655, miR-431, miR-891a, and miR-887 were associated with oligometastatic disease. MiR-205, miR-216b, and miR-506 were associated with polymetastatic disease.
Full table
The immune microenvironment
Innate and adaptive immune cells in the lung tumor microenvironment harbor both tumor-promoting and tumor-suppressing activities, and the interaction between the two predicts clinical outcome (128,129). To understand the immune signature in lung adenocarcinoma, Kim et al. performed comparative analysis between normal epithelial and tumor cells, and between primary tumor and metastatic foci from surgical resection samples (130). Primary tumors were enriched with T lymphocytes and myeloid cells, indicating the activation of adaptive immune responses. Myeloid cells were abundant in metastatic lymph nodes compared to normal lymph nodes indicating an association of myeloid infiltration with metastasis (Figure 2). In both primary tumor and metastatic sites, there was a simultaneous decrease of regulatory T cells and an increase in the proportions of plasmacytoid dendritic cells, creating an immunosuppressive microenvironment with sub-optimal tumor antigen presentation (130,131). Exhausted CD8+ T cells and monocyte-derived macrophages were increased in the metastatic lesions and metastatic lymph nodes. The relative proportion of B cells was increased in primary tumors, compared to the normal lung tissue, suggesting highly activated humoral immune responses in some lung adenocarcinoma patients. Genome-wide expression analysis has begun to provide molecular insights into this tumor-induced reprogramming of infiltrating lymphoid and myeloid cells, with myeloid cells from tumors and matched adjacent non-neoplastic lung tissue exhibiting differentially regulated genes (132,133). Among them, expression of gene encoding osteopontin (OPN), a secreted phosphoglycoprotein that has been shown to contribute to tumor progression and metastasis, was >1,000-fold upregulated in intra-tumoral myeloid cells (134). Thrombospondin-1 (TSP1), an anti-tumorigenic factor that inhibits angiogenesis by reducing endothelial cell migration and survival, is downregulated in intra-tumoral myeloid cells, which promotes NSCLC growth (133,135). Studies have also shown discordance in PD-L1 expression in the tumor cells of NSCLC between the primary and metastatic sites (136,137).
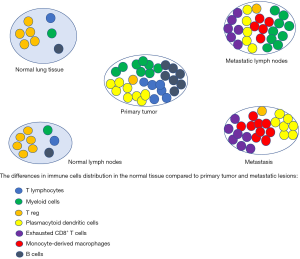
Further evidence on how the immune signature controls tumor progression comes from a study done by Pitroda et al., where colorectal cancer samples were divided into three cohorts based on immune signaling. Among these subtypes, the immune enriched subtype was associated with limited metastases and better outcome (27). Van den Eynde et al. predicted risk of recurrence using the “Immunoscore”, which is a representation of T cell infiltration of colorectal tumor specimens (138). Patients with a high Immunoscore (representative of high immune infiltrate) and 1 to 3 metastases had a significantly better outcome than patients with either a high Immunoscore and ≥4 metastases or a low Immunoscore.
Biomarkers to identify oligometastatic disease
Circulating tumor DNA (ctDNA)
Often referred to as “liquid biopsy”, it is now possible to detect double-stranded DNA fragments released from tumor cells into the circulation during apoptosis or the necrotic process (variably termed ctDNA or cell-free DNA). This technology appears potentially useful for distinguishing oligometastatic from polymetastatic disease. On a molecular level, ctDNA carries important genomic information from the total burden of the tumor, which may help identify dynamic changes that occur in metastatic tumor progression (139). For example, in pancreatic cancer ctDNA KRAS mutations are identified more frequently in patients with metastatic disease and are associated with poor survival (140). Alterations in RAS, BRAF, ERBB2 genes detected through ctDNA are more frequently observed in colorectal cancer patients with high tumor burden (141). Compared to other serum biomarkers, such as carcinoembryonic antigen (CEA), and carbohydrate antigen 19-9 (CA19-9), these ctDNA alterations occurred earlier and had greater sensitivity and specificity (140). ctDNA may also provide information regarding epigenetic modifications, including methylation of specific genes promoting metastasis development (141). For example, loss of 5-hydroxymethylcytosine (5hmc), a DNA pyrimidine nitrogen base with epigenetic functions, in lung tumor DNA is associated with progression into metastatic disease (142).
Separately, the concentration of ctDNA in plasma correlates with tumor burden. Patients with higher concentrations of ctDNA in plasma are more likely to have higher tumor burden and distant disease (143). Using ctDNA assays in oligometastatic disease in patients that are candidates for local consolidative therapy in the pre-treatment and post-treatment setting improves the accuracy of early detection of metastatic disease and disease recurrence (144). By measuring circulating tumor specific biomarkers, ctDNA and/or ctRNA assays may also facilitate discovery of prospective biomarkers for patients who are most likely to benefit from local consolidative therapy, as obtaining a tissue biopsy in these patients can often be challenging. A key limitation relevant to oligometastatic disease, the lower the burden of the disease, the less sensitive ctDNA profiling becomes and more genomic alterations are missed (143).
Cytokines
Cytokines play indispensable roles in inflammation and antitumor immune response. Tang et al. investigated the peripheral blood cytokines and correlated the findings with survival in NSCLC (144). The results showed that certain peripheral cytokines—such as IL-1α, which is known to initiate an inflammatory cascade that facilitates neutrophil mobilization and antitumor activity—are associated with survival (144,145). Several proangiogenic cytokines are also present at the tumor site such as: platelet-derived growth factor (PDGF), fibroblast growth factor (FGF)-2, FGF-6, IL-6, IL-8, vascular endothelial growth factor (VEGF) and angiopoietin which are responsible for promoting tumor growth and increasing tumor blood vessel density (146). NSCLC tumor cells secretes IL-17 which in turn attracts tumor associated macrophages. Tumor associated macrophages secretes cyclogenase-2 (COX2), matrix metalloproteinase-9 (MMP9), PDGF-B, VEGFA, hepatocyte growth factor (HGF), cathepsin-k to increase tumor invasiveness (147). Adenocarcinoma-associated CAFs also secrete immunomodulatory cytokines such as transforming growth factor β (TGF-β) and VEGF inducing forkhead box P3 expressing regulatory T-cells that are correlated with a poor outcome in lung adenocarcinoma (148). Moving forward, cytokine analysis may be used to determine the potential for progression into polymetastatic disease (144).
Circulating tumor cells (CTCs)
The detection of tumor cells that shed from the tumor to the circulation correlates with prognosis and therapeutic efficacy (149-151). Termed CTCs, they may also be used to monitor for postsurgical cancer relapse (152). Some studies have used identification of aneuploid CTCs (apCTCs) as a marker to predict oligometastatic disease (153,154). Circulating tumor endothelial cells (CTECs) may provide additional information regarding the cancer invasiveness, metastatic potential, and progression (155,156). In addition, cancer associated fibroblasts have been proposed to circulate together with CTCs to help support cancer metastasis. A strong reciprocal interaction between the CTCs and the blood microenvironment including the platelets and the neutrophils has been reported. The CTCs activate and educate the platelets, while the platelets protect the CTCs (157). The ATP released through CTC-induced platelet aggregation binds to the P2Y2 receptor, stimulating intravasation and metastases development (158). Furthermore, the adherence of platelets at the surface of CTCs protects the CTCs from being recognized by the immune cells thereby promotes CTCs survival (159). Blocking this interaction using P2Y12 inhibitor (ticagrelor) or aspirin, has been studied as a tool to reduce metastases (160,161). Another important interaction is between the neutrophils and the CTCs. The neutrophils generate neutrophil extracellular traps by secreting their chromatin content (162). While this process was initially thought to be a mechanism to kill bacteria, recent reports show that this mechanism promotes metastases though increased migration and proliferation of CTCs (163).
Similarity network fusions (SNFs)
SNF is a new computational method for data integration. Briefly, it entails constructing networks of samples (e.g., patients) for each available data type (such as mRNA expression data, DNA methylation, clinical data, questionnaires, imaging data, etc.), and then efficiently fusing these into one network that represents the full spectrum of underlying data. SNF excels over single data type analysis and established integrative approaches when identifying cancer subtypes as it reduces collection bias and noise in different data types and is effective for predicting survival. Data on the correlation of SNFs and metastatic virulence in lung cancer are lacking. However, certain SNFs were found to be associated with distinct molecular subtypes as well as distinct clinical outcomes in other malignancies (27). For example, using SNFs, Pitroda et al. stratified patients with colorectal carcinoma with limited liver metastases into three distinct molecular subtypes: (I) canonical (II) immune and (III) stromal (27). The immune subtype was associated with lower recurrence rate and longer survival after hepatic metastasectomy, while the canonical and the stromal subtypes were associated with poor survival. Furthermore, integration of the resultant molecular subtypes with clinical risk stratification yielded three prognostic risk groups: low-risk, intermediate-risk, and high-risk. The integrated low-risk group showed significantly longer distant metastasis-free survival and overall survival and was largely consistent with the oligometastatic phenotype.
The concept of treating oligoprogression
Differences between primary and metastatic cancer lesions
Most research into genomic and molecular characteristics of cancer is based on primary tumor samples, even in cases of metastatic disease. As a result, molecular features of metastatic sites and their association with clinical outcomes represents an understudied area. In lung cancer, driver mutations are classified into trunk (initiating) mutations and branching mutations (164,165). While trunk driver mutations initiate the formation of the primary tumor, branching driver mutations lead to subclonal evolution of the malignancy. Most activating mutations in the EGFR, BRAF, KRAS, MET, RET, ROS1 and ALK are trunk drivers (165) and are commonly concordant in primary and metastatic tumors. However, many reports have shown discordant trunk mutations between paired primary and metastatic lung cancer specimens suggesting the presence of tumor heterogeneity (166). Different metastatic sites from the same patient may feature different genomic and epigenetic signatures and thus can have different malignant potential (12-14,164,167-171). Furthermore, within a primary tumor, cells may have varying metastatic potential (172,173). In lung adenocarcinoma, it has been shown that lymph node metastases have greater expression of ALK (8% vs. 1%), EGFR (50% vs. 42%), PD-L1 (36% vs. 25%) and ROS-1 (3% vs. 1%) compared to primary tumors. Distant organ metastases also exhibited higher cMET amplification (7% vs. 3%) than primary tumors. Similarly, squamous carcinomas showed higher ALK expression (10% vs. 1%) and PD-L1 expression (42% vs. 33%) in lymph node metastases compared to the primary tumor (33%) (136). It has been suggested that oligo- or poly-metastases may either originate from different clones or may be part of a sequential development, with oligometastasis representing a transient state in the metastatic process (56).
The approach to molecularly targetable oligometastatic NSCLC
The current gold standard to identify the oligometastatic state is to determine the number of metastatic sites evident on conventional imaging. However, this definition does not account for tumor markers and genomic signatures, which may strongly influence survival. The list of molecularly defined subtypes of NSCLC—which have distinct prognosis and treatment—continues to expand. While more data are needed regarding the differences in clinical outcomes between molecularly targetable oligometastatic NSCLC and non-targetable oligometastatic NSCLC; it is clear that these clinical entities require different therapeutic approaches. Incorporation of EGFR mutation status in advanced NSCLC further differentiates survival curves in the metastatic setting and predicts survival more precisely than the number of metastatic sites (174). Patients with oligometastatic disease who harbor the EGFR mutation and receive molecularly targeted therapies have superior outcomes compared to oligometastatic patients without the EGFR mutation (175).
Treating oligoprogression in molecularly targetable oligometastatic NSCLC
Although patients with NSCLC harboring driver mutations have high rates of response to tyrosine kinase inhibitors (TKIs), depending on the molecular target resistance generally develops after 10–20 months of treatment, even with use of state-of-the-art third generation TKIs such as osimertinib (176). One of the frequently seen situations when treating these patients is the progression of a single or few clinically detectable metastatic lesions while other metastases respond to treatment. This represents the “escape” of a resistant subclone that drives progression. This suggests that future diagnostic and therapeutic decision-making will need to be based on tissue retrieved directly from metastatic tissue rather than inferred from previously resected primary tumor. Several studies have shown that aggressive localized management of these resistant subclones may preserve the efficacy of a relatively nontoxic systemic treatment and leave the patient with more options over time (177,178).
Conclusions
The current approach to identifying oligometastatic disease incorporates baseline imaging characteristics and clinical behavior, while reserving aggressive, local treatment of both primary and oligometastatic site(s) until after 6–12 months of systemic therapy designed to allow the natural history of disease to declare itself, such approaches are far from ideal.
Earlier understanding of a true oligometastatic state, through integration of molecular prognostic classifiers such as SNFs, ctDNA, CTCs and cytokines along with other clinical features, might allow earlier and more effective use of local therapies. It would also enhance understanding and reduce uncertainty among patients. Although still largely exploratory, incorporating molecular characteristics and biomarkers with clinical features and conventional imaging studies appears highly promising for improving the accuracy of defining and classifying oligometastatic NSCLC. Such advances may decrease morbidity and cost by eliminating futile localized therapies, improve the efficacy of indicated localized therapies, and ultimately enhance the quality and quantity of life for patients with lung cancer.
Acknowledgments
Funding: This study was supported in part by the University of Texas Specialized Program in Research Excellence (SPORE, P50CA070907-21) and a National Cancer Institute Midcareer Award in Patient-Oriented Research (K24CA201543-01).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Maurizio Infante & Thierry Berghmans) for the series “Oligometastatic NSCLC: definition and treatment opportunities” published in Translational Lung Cancer Research. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-448
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-21-448
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-448). The series “Oligometastatic NSCLC: definition and treatment opportunities” was commissioned by the editorial office without any funding or sponsorship. JDM serves as an unpaid editorial board member of Translational Lung Cancer Research. JDM receives royalties/licensing from the NIH and UTSW for distribution of tumor cell lines. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hendriks LE, Dingemans AM, De Ruysscher D. Proposals for the M-descriptors of the Eight TNM Classification for Non-Small Cell Lung Cancer: Are More Data Needed? J Thorac Oncol 2016;11:e42-3. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aggarwal C, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 1.2020. J Natl Compr Canc Netw 2019;17:1464-72. [Crossref] [PubMed]
- Gomez DR, Blumenschein GR Jr, Lee JJ, et al. Local consolidative therapy versus maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer without progression after first-line systemic therapy: a multicentre, randomised, controlled, phase 2 study. Lancet Oncol 2016;17:1672-82. [Crossref] [PubMed]
- Iyengar P, Kavanagh BD, Wardak Z, et al. Phase II trial of stereotactic body radiation therapy combined with erlotinib for patients with limited but progressive metastatic non-small-cell lung cancer. J Clin Oncol 2014;32:3824-30. [Crossref] [PubMed]
- Casiraghi M, Bertolaccini L, Sedda G, et al. Lung cancer surgery in oligometastatic patients: outcome and survival. Eur J Cardiothorac Surg 2020;57:1173-80. [Crossref] [PubMed]
- Halsted WS. I. The Results of Radical Operations for the Cure of Carcinoma of the Breast. Ann Surg 1907;46:1-19. [Crossref] [PubMed]
- Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev 1989;8:98-101. [PubMed]
- Weichselbaum RR, Hellman S. Oligometastases revisited. Nat Rev Clin Oncol 2011;8:378-82. [Crossref] [PubMed]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563-72. [Crossref] [PubMed]
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer 2003;3:453-8. [Crossref] [PubMed]
- Corbin KS, Hellman S, Weichselbaum RR. Extracranial oligometastases: a subset of metastases curable with stereotactic radiotherapy. J Clin Oncol 2013;31:1384-90. [Crossref] [PubMed]
- Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet 2003;33:49-54. [Crossref] [PubMed]
- Dhanasekaran SM, Barrette TR, Ghosh D, et al. Delineation of prognostic biomarkers in prostate cancer. Nature 2001;412:822-6. [Crossref] [PubMed]
- Haffner MC, Mosbruger T, Esopi DM, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest 2013;123:4918-22. [Crossref] [PubMed]
- Foster CC, Pitroda SP, Weichselbaum RR. Definition, Biology, and History of Oligometastatic and Oligoprogressive Disease. Cancer J 2020;26:96-9. [Crossref] [PubMed]
- Reyes DK, Pienta KJ. The biology and treatment of oligometastatic cancer. Oncotarget 2015;6:8491-524. [Crossref] [PubMed]
- Bergsma DP, Salama JK, Singh DP, et al. Radiotherapy for Oligometastatic Lung Cancer. Front Oncol 2017;7:210. [Crossref] [PubMed]
- Giaj-Levra N, Giaj Levra M, Berghmans T, et al. Oligometastatic non-small cell lung cancer (NSCLC): Does number of metastasis matter? Lung Cancer 2020;139:216-8. [Crossref] [PubMed]
- Ashworth AB, Senan S, Palma DA, et al. An individual patient data metaanalysis of outcomes and prognostic factors after treatment of oligometastatic non-small-cell lung cancer. Clin Lung Cancer 2014;15:346-55. [Crossref] [PubMed]
- Opitz I, Patella M, Payrard L, et al. Prognostic factors of oligometastatic non-small-cell lung cancer following radical therapy: a multicentre analysis. Eur J Cardiothorac Surg 2020;57:1166-72. [Crossref] [PubMed]
- Gomez DR, Tang C, Zhang J, et al. Local Consolidative Therapy Vs. Maintenance Therapy or Observation for Patients With Oligometastatic Non-Small-Cell Lung Cancer: Long-Term Results of a Multi-Institutional, Phase II, Randomized Study. J Clin Oncol 2019;37:1558-65. [Crossref] [PubMed]
- Mitchell KG, Farooqi A, Ludmir EB, et al. Improved Overall Survival With Comprehensive Local Consolidative Therapy in Synchronous Oligometastatic Non-Small-Cell Lung Cancer. Clin Lung Cancer 2020;21:37-46.e7. [Crossref] [PubMed]
- Pastorino U, Buyse M, Friedel G, et al. Long-term results of lung metastasectomy: prognostic analyses based on 5206 cases. J Thorac Cardiovasc Surg 1997;113:37-49. [Crossref] [PubMed]
- Lussier YA, Khodarev NN, Regan K, et al. Oligo- and polymetastatic progression in lung metastasis(es) patients is associated with specific microRNAs. PLoS One 2012;7:e50141 [Crossref] [PubMed]
- Gutiontov SI, Pitroda SP, Weichselbaum RR. Oligometastasis: Past, Present, Future. Int J Radiat Oncol Biol Phys 2020;108:530-8. [Crossref] [PubMed]
- Lo SS, Moffatt-Bruce SD, Dawson LA, et al. The role of local therapy in the management of lung and liver oligometastases. Nat Rev Clin Oncol 2011;8:405-16. [Crossref] [PubMed]
- Pitroda SP, Khodarev NN, Huang L, et al. Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat Commun 2018;9:1793. [Crossref] [PubMed]
- Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol 2015;10:431-7. [Crossref] [PubMed]
- Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012;30:2956-62. [Crossref] [PubMed]
- Stremitzer S, Stift J, Gruenberger B, et al. KRAS status and outcome of liver resection after neoadjuvant chemotherapy including bevacizumab. Br J Surg 2012;99:1575-82. [Crossref] [PubMed]
- Bruin SC, de Ronde JJ, Wiering B, et al. Selection of patients for hepatic surgery of colorectal cancer liver metastasis based on genomic aberrations. Ann Surg Oncol 2013;20:S560-9. [Crossref] [PubMed]
- Wang Y, Xue Q, Zheng Q, et al. SMAD4 mutation correlates with poor prognosis in non-small cell lung cancer. Lab Invest 2021;101:463-76. [Crossref] [PubMed]
- Hassan WA, Yoshida R, Kudoh S, et al. Notch1 controls cell invasion and metastasis in small cell lung carcinoma cell lines. Lung Cancer 2014;86:304-10. [Crossref] [PubMed]
- Uppal A, Wightman SC, Mallon S, et al. 14q32-encoded microRNAs mediate an oligometastatic phenotype. Oncotarget 2015;6:3540-52. [Crossref] [PubMed]
- Desai A, Adjei AA. FGFR Signaling as a Target for Lung Cancer Therapy. J Thorac Oncol 2016;11:9-20. [Crossref] [PubMed]
- Burbee DG, Forgacs E, Zöchbauer-Müller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 2001;93:691-9. [Crossref] [PubMed]
- Yoo HC, Park SJ, Nam M, et al. A Variant of SLC1A5 Is a Mitochondrial Glutamine Transporter for Metabolic Reprogramming in Cancer Cells. Cell Metab 2020;31:267-283.e12. [Crossref] [PubMed]
- Elia I, Doglioni G, Fendt SM. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol 2018;28:673-84. [Crossref] [PubMed]
- Jiang Y, Pan Y, Rhea PR, et al. A Sucrose-Enriched Diet Promotes Tumorigenesis in Mammary Gland in Part through the 12-Lipoxygenase Pathway. Cancer Res 2016;76:24-9. [Crossref] [PubMed]
- Mashimo T, Pichumani K, Vemireddy V, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 2014;159:1603-14. [Crossref] [PubMed]
- Liu Y, Metzinger MN, Lewellen KA, et al. Obesity Contributes to Ovarian Cancer Metastatic Success through Increased Lipogenesis, Enhanced Vascularity, and Decreased Infiltration of M1 Macrophages. Cancer Res 2015;75:5046-57. [Crossref] [PubMed]
- Chen M, Zhang J, Sampieri K, et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet 2018;50:206-18. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Bu P, Chen KY, Xiang K, et al. Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab 2018;27:1249-1262.e4. [Crossref] [PubMed]
- Menard JA, Christianson HC, Kucharzewska P, et al. Metastasis Stimulation by Hypoxia and Acidosis-Induced Extracellular Lipid Uptake Is Mediated by Proteoglycan-Dependent Endocytosis. Cancer Res 2016;76:4828-40. [Crossref] [PubMed]
- Fainsod-Levi T, Gershkovitz M, Völs S, et al. Hyperglycemia Impairs Neutrophil Mobilization Leading to Enhanced Metastatic Seeding. Cell Rep 2017;21:2384-92. [Crossref] [PubMed]
- Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016;16:582-98. [Crossref] [PubMed]
- Wang L, Cao L, Wang H, et al. Cancer-associated fibroblasts enhance metastatic potential of lung cancer cells through IL-6/STAT3 signaling pathway. Oncotarget 2017;8:76116-28. [Crossref] [PubMed]
- Navab R, Strumpf D, To C, et al. Integrin α11β1 regulates cancer stromal stiffness and promotes tumorigenicity and metastasis in non-small cell lung cancer. Oncogene 2016;35:1899-908. [Crossref] [PubMed]
- Lakins MA, Ghorani E, Munir H, et al. Cancer-associated fibroblasts induce antigen-specific deletion of CD8 + T Cells to protect tumour cells. Nat Commun 2018;9:948. [Crossref] [PubMed]
- Cheng HW, Onder L, Cupovic J, et al. CCL19-producing fibroblastic stromal cells restrain lung carcinoma growth by promoting local antitumor T-cell responses. J Allergy Clin Immunol 2018;142:1257-1271.e4. [Crossref] [PubMed]
- Ao Z, Shah SH, Machlin LM, et al. Identification of Cancer-Associated Fibroblasts in Circulating Blood from Patients with Metastatic Breast Cancer. Cancer Res 2015;75:4681-7. [Crossref] [PubMed]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597-610. [Crossref] [PubMed]
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593-601. [Crossref] [PubMed]
- Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 2009;23:2140-51. [Crossref] [PubMed]
- Uppal A, Ferguson MK, Posner MC, et al. Towards a molecular basis of oligometastatic disease: potential role of micro-RNAs. Clin Exp Metastasis 2014;31:735-48. [Crossref] [PubMed]
- Lussier YA, Xing HR, Salama JK, et al. MicroRNA expression characterizes oligometastasis(es). PLoS One 2011;6:e28650 [Crossref] [PubMed]
- Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008;13:48-57. [Crossref] [PubMed]
- Aiso T, Ohtsuka K, Ueda M, et al. Serum levels of candidate microRNA diagnostic markers differ among the stages of non-small-cell lung cancer. Oncol Lett 2018;16:6643-51. [Crossref] [PubMed]
- Li J, Yang S, Yan W, et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest 2015;95:1056-70. [Crossref] [PubMed]
- Edme N, Downward J, Thiery JP, et al. Ras induces NBT-II epithelial cell scattering through the coordinate activities of Rac and MAPK pathways. J Cell Sci 2002;115:2591-601. [Crossref] [PubMed]
- Liu B, Wu X, Liu B, et al. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta 2012;1822:1692-704. [Crossref] [PubMed]
- Zhou H, Huang Z, Chen X, et al. miR-98 inhibits expression of TWIST to prevent progression of non-small cell lung cancers. Biomed Pharmacother 2017;89:1453-61. [Crossref] [PubMed]
- Jin X, Yu Y, Zou Q, et al. MicroRNA-105 promotes epithelial-mesenchymal transition of nonsmall lung cancer cells through upregulating Mcl-1. J Cell Biochem 2019;120:5880-8. [Crossref] [PubMed]
- Kim YS, Yi BR, Kim NH, et al. Role of the epithelial-mesenchymal transition and its effects on embryonic stem cells. Exp Mol Med 2014;46:e108 [Crossref] [PubMed]
- Lin CW, Chang YL, Chang YC, et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun 2013;4:1877. [Crossref] [PubMed]
- Chang TH, Tsai MF, Gow CH, et al. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett 2017;402:190-202. [Crossref] [PubMed]
- Li H, Ouyang R, Wang Z, et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep 2016;6:39001. [Crossref] [PubMed]
- Liu XH, Lu KH, Wang KM, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer 2012;12:348. [Crossref] [PubMed]
- Larzabal L, de Aberasturi AL, Redrado M, et al. TMPRSS4 regulates levels of integrin α5 in NSCLC through miR-205 activity to promote metastasis. Br J Cancer 2014;110:764-74. [Crossref] [PubMed]
- Wang Z, Gao S, Li T, et al. Exploring the microRNA profiles as potential diagnostic probes for oligo- and polymetastatic prognosis of lung metastasis(es) patients. Medicine (Baltimore) 2018;97:e10958 [Crossref] [PubMed]
- Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009;16:498-509. [Crossref] [PubMed]
- Arora S, Ranade AR, Tran NL, et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer 2011;129:2621-31. [Crossref] [PubMed]
- Sun CC, Li SJ, Yuan ZP, et al. MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway. Aging (Albany NY) 2016;8:2509-24. [Crossref] [PubMed]
- Wang J, Wang Y, Sun D, et al. miR-455-5p promotes cell growth and invasion by targeting SOCO3 in non-small cell lung cancer. Oncotarget 2017;8:114956-65. [Crossref] [PubMed]
- Mo X, Zhang F, Liang H, et al. miR-544a promotes the invasion of lung cancer cells by targeting cadherina 1 in vitro. Onco Targets Ther 2014;7:895-900. [Crossref] [PubMed]
- Liu Y, Wang F, Xu P. miR-590 accelerates lung adenocarcinoma migration and invasion through directly suppressing functional target OLFM4. Biomed Pharmacother 2017;86:466-74. [Crossref] [PubMed]
- Zhu X, Ju S, Yuan F, et al. microRNA-664 enhances proliferation, migration and invasion of lung cancer cells. Exp Ther Med 2017;13:3555-62. [Crossref] [PubMed]
- Tominaga E, Yuasa K, Shimazaki S, et al. MicroRNA-1 targets Slug and endows lung cancer A549 cells with epithelial and anti-tumorigenic properties. Exp Cell Res 2013;319:77-88. [Crossref] [PubMed]
- Zhang K, Li XY, Wang ZM, et al. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci 2017;21:3598-604. [PubMed]
- Jiang BY, Zhang XC, Su J, et al. BCL11A overexpression predicts survival and relapse in non-small cell lung cancer and is modulated by microRNA-30a and gene amplification. Mol Cancer 2013;12:61. [Crossref] [PubMed]
- Yang L, Yang J, Li J, et al. MircoRNA-33a inhibits epithelial-to-mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer. Sci Rep 2015;5:13677. [Crossref] [PubMed]
- Qu J, Li M, An J, et al. MicroRNA-33b inhibits lung adenocarcinoma cell growth, invasion, and epithelial-mesenchymal transition by suppressing Wnt/β-catenin/ZEB1 signaling. Int J Oncol 2015;47:2141-52. [Crossref] [PubMed]
- Ahn YH, Gibbons DL, Chakravarti D, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest 2012;122:3170-83. [Crossref] [PubMed]
- Liu X, Tian XD, Liu Y, et al. Regulation of Twist in the metastasis of non-small cell lung cancer by miR-92b. Eur Rev Med Pharmacol Sci 2016;20:4003-10. [PubMed]
- Han L, Chen W, Xia Y, et al. MiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1. Am J Transl Res 2018;10:1172-83. [PubMed]
- Li Z, Wang X, Li W, et al. miRNA-124 modulates lung carcinoma cell migration and invasion. Int J Clin Pharmacol Ther 2016;54:603-12. [Crossref] [PubMed]
- Jia Z, Zhang Y, Xu Q, et al. miR-126 suppresses epithelial-to-mesenchymal transition and metastasis by targeting PI3K/AKT/Snail signaling of lung cancer cells. Oncol Lett 2018;15:7369-75. [Crossref] [PubMed]
- Hu J, Cheng Y, Li Y, et al. microRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. Eur J Cancer 2014;50:2336-50. [Crossref] [PubMed]
- You J, Li Y, Fang N, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One 2014;9:e91827 [Crossref] [PubMed]
- Yang ZQ, Wu CA, Cheng YX. Prognostic Value of microRNA-133a Expression and Its Clinicopathologic Significance in Non-Small Cell Lung Cancer: A Comprehensive Study Based on Meta-Analysis and the TCGA Database. Oncol Res Treat 2018;41:762-8. [Crossref] [PubMed]
- Yang Y, Liu L, Cai J, et al. Targeting Smad2 and Smad3 by miR-136 suppresses metastasis-associated traits of lung adenocarcinoma cells. Oncol Res 2013;21:345-52. [Crossref] [PubMed]
- Jin Z, Guan L, Song Y, et al. MicroRNA-138 regulates chemoresistance in human non-small cell lung cancer via epithelial mesenchymal transition. Eur Rev Med Pharmacol Sci 2016;20:1080-6. [PubMed]
- Zhang G, An H, Fang X. MicroRNA-144 regulates proliferation, invasion, and apoptosis of cells in malignant solitary pulmonary nodule via zinc finger E-box-binding homeobox 1. Int J Clin Exp Pathol 2015;8:5960-7. [PubMed]
- Liu Q, Chen J, Wang B, et al. miR-145 modulates epithelial-mesenchymal transition and invasion by targeting ZEB2 in non-small cell lung cancer cell lines. J Cell Biochem 2018; Epub ahead of print. [Crossref] [PubMed]
- Li J, Song Y, Wang Y, et al. MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem 2013;380:277-82. [Crossref] [PubMed]
- Luo H, Liang C. MicroRNA-148b inhibits proliferation and the epithelial-mesenchymal transition and increases radiosensitivity in non-small cell lung carcinomas by regulating ROCK1. Exp Ther Med 2018;15:3609-16. [Crossref] [PubMed]
- Lin X, Yang Z, Zhang P, et al. miR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncol Lett 2016;12:301-6. [Crossref] [PubMed]
- Lin J, Chen Y, Liu L, et al. MicroRNA-155-5p suppresses the migration and invasion of lung adenocarcinoma A549 cells by targeting Smad2. Oncol Lett 2018;16:2444-52. [Crossref] [PubMed]
- Yang J, Liu H, Wang H, et al. Down-regulation of microRNA-181b is a potential prognostic marker of non-small cell lung cancer. Pathol Res Pract 2013;209:490-4. [Crossref] [PubMed]
- Yang CL, Zheng XL, Ye K, et al. MicroRNA-183 Acts as a Tumor Suppressor in Human Non-Small Cell Lung Cancer by Down-Regulating MTA1. Cell Physiol Biochem 2018;46:93-106. [Crossref] [PubMed]
- Xu W, Luo F, Sun B, et al. HIF-2α, acting via miR-191, is involved in angiogenesis and metastasis of arsenite-transformed HBE cells. Toxicol Res (Camb) 2016;5:66-78. [Crossref] [PubMed]
- Yongchun Z, Linwei T, Xicai W, et al. MicroRNA-195 inhibits non-small cell lung cancer cell proliferation, migration and invasion by targeting MYB. Cancer Lett 2014;347:65-74. [Crossref] [PubMed]
- Diepenbruck M, Tiede S, Saxena M, et al. miR-1199-5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nat Commun 2017;8:1168. [Crossref] [PubMed]
- Wang J, Zhou F, Yin L, et al. MicroRNA-199b targets the regulation of ZEB1 expression to inhibit cell proliferation, migration and invasion in non-small cell lung cancer. Mol Med Rep 2017;16:5007-14. [Crossref] [PubMed]
- Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008;9:582-9. [Crossref] [PubMed]
- Zeng Y, Zhu J, Shen D, et al. Repression of Smad4 by miR-205 moderates TGF-β-induced epithelial-mesenchymal transition in A549 cell lines. Int J Oncol 2016;49:700-8. [Crossref] [PubMed]
- Chen QY, Jiao DM, Wang J, et al. miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget 2016;7:24510-26. [Crossref] [PubMed]
- Hou Y, Zhen J, Xu X, et al. miR-215 functions as a tumor suppressor and directly targets ZEB2 in human non-small cell lung cancer. Oncol Lett 2015;10:1985-92. [Crossref] [PubMed]
- Wang RT, Xu M, Xu CX, et al. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin Cancer Res 2014;20:4705-16. [Crossref] [PubMed]
- Shi ZM, Wang L, Shen H, et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene 2017;36:2577-88. [Crossref] [PubMed]
- Li Q, Ran P, Zhang X, et al. Downregulation of N-Acetylglucosaminyltransferase GCNT3 by miR-302b-3p Decreases Non-Small Cell Lung Cancer (NSCLC) Cell Proliferation, Migration and Invasion. Cell Physiol Biochem 2018;50:987-1004. [Crossref] [PubMed]
- Ma W, Ma CN, Zhou NN, et al. Up- regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Sci Rep 2016;6:31651. [Crossref] [PubMed]
- Chen W, Wang J, Liu S, et al. MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J Exp Clin Cancer Res 2016;35:76. [Crossref] [PubMed]
- Hu WW, Chen PC, Chen JM, et al. Periostin promotes epithelial-mesenchymal transition via the MAPK/miR-381 axis in lung cancer. Oncotarget 2017;8:62248-60. [Crossref] [PubMed]
- Shan C, Fei F, Li F, et al. miR-448 is a novel prognostic factor of lung squamous cell carcinoma and regulates cells growth and metastasis by targeting DCLK1. Biomed Pharmacother 2017;89:1227-34. [Crossref] [PubMed]
- He Z, Xia Y, Pan C, et al. Up-Regulation of MiR-452 Inhibits Metastasis of Non-Small Cell Lung Cancer by Regulating BMI1. Cell Physiol Biochem 2015;37:387-98. [Crossref] [PubMed]
- Li YJ, Ping C, Tang J, et al. MicroRNA-455 suppresses non-small cell lung cancer through targeting ZEB1. Cell Biol Int 2016;40:621-8. [Crossref] [PubMed]
- Huang RS, Zheng YL, Li C, et al. MicroRNA-485-5p suppresses growth and metastasis in non-small cell lung cancer cells by targeting IGF2BP2. Life Sci 2018;199:104-11. [Crossref] [PubMed]
- Gong F, Ren P, Zhang Y, et al. MicroRNAs-491-5p suppresses cell proliferation and invasion by inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res 2016;8:485-95. [PubMed]
- Yin Q, Han Y, Zhu D, et al. miR-145 and miR-497 suppress TGF-β-induced epithelial-mesenchymal transition of non-small cell lung cancer by targeting MTDH. Cancer Cell Int 2018;18:105. [Crossref] [PubMed]
- Guo S, Yang P, Jiang X, et al. Genetic and epigenetic silencing of mircoRNA-506-3p enhances COTL1 oncogene expression to foster non-small lung cancer progression. Oncotarget 2017;8:644-57. [Crossref] [PubMed]
- Zhang L, Yu S. Role of miR-520b in non-small cell lung cancer. Exp Ther Med 2018;16:3987-95. [Crossref] [PubMed]
- Wang FF, Wang S, Xue WH, et al. microRNA-590 suppresses the tumorigenesis and invasiveness of non-small cell lung cancer cells by targeting ADAM9. Mol Cell Biochem 2016;423:29-37. [Crossref] [PubMed]
- Tong X, Su P, Yang H, et al. MicroRNA-598 inhibits the proliferation and invasion of non-small cell lung cancer cells by directly targeting ZEB2. Exp Ther Med 2018;16:5417-23. [Crossref] [PubMed]
- Xu L, Xu X, Huang H, et al. MiR-1260b promotes the migration and invasion in non-small cell lung cancer via targeting PTPRK. Pathol Res Pract 2018;214:776-83. [Crossref] [PubMed]
- Capodanno A, Boldrini L, Proietti A, et al. Let-7g and miR-21 expression in non-small cell lung cancer: correlation with clinicopathological and molecular features. Int J Oncol 2013;43:765-74. [Crossref] [PubMed]
- Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717-34. [Crossref] [PubMed]
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014-22. [Crossref] [PubMed]
- Kim N, Kim HK, Lee K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020;11:2285. [Crossref] [PubMed]
- Demoulin S, Herfs M, Delvenne P, et al. Tumor microenvironment converts plasmacytoid dendritic cells into immunosuppressive/tolerogenic cells: insight into the molecular mechanisms. J Leukoc Biol 2013;93:343-52. [Crossref] [PubMed]
- Lavín JL, Sánchez-Morán M, Bárcena L, et al. A fistful of tips for a fruitful high throughput sequencing experiment. Bioessays 2017; [Crossref] [PubMed]
- Durrans A, Gao D, Gupta R, et al. Identification of Reprogrammed Myeloid Cell Transcriptomes in NSCLC. PLoS One 2015;10:e0129123 [Crossref] [PubMed]
- Rittling SR, Chambers AF. Role of osteopontin in tumour progression. Br J Cancer 2004;90:1877-81. [Crossref] [PubMed]
- Lawler J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J Cell Mol Med 2002;6:1-12. [Crossref] [PubMed]
- Gatalica Z, Feldman R, Russell K, et al. 1PD Differences in expression of predictive biomarkers between primary and metastatic non-small cell lung cancer tumors. J Thorac Oncol 2016;11:S57-67. [Crossref]
- Senarathne W, Vranic S, Xiu J, et al. Composition of the immune microenvironment differs between carcinomas metastatic to the lungs and primary lung carcinomas. Ann Diagn Pathol 2018;33:62-8. [Crossref] [PubMed]
- Van den Eynde M, Mlecnik B, Bindea G, et al. The Link between the Multiverse of Immune Microenvironments in Metastases and the Survival of Colorectal Cancer Patients. Cancer Cell 2018;34:1012-1026.e3. [Crossref] [PubMed]
- Namløs HM, Boye K, Mishkin SJ, et al. Noninvasive Detection of ctDNA Reveals Intratumor Heterogeneity and Is Associated with Tumor Burden in Gastrointestinal Stromal Tumor. Mol Cancer Ther 2018;17:2473-80. [Crossref] [PubMed]
- Bernard V, Kim DU, San Lucas FA, et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019;156:108-118.e4. [Crossref] [PubMed]
- Germano G, Mauri G, Siravegna G, et al. Parallel Evaluation of Circulating Tumor DNA and Circulating Tumor Cells in Metastatic Colorectal Cancer. Clin Colorectal Cancer 2018;17:80-3. [Crossref] [PubMed]
- Song CX, Yin S, Ma L, et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 2017;27:1231-42. [Crossref] [PubMed]
- Strickler JH, Loree JM, Ahronian LG, et al. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov 2018;8:164-73. [Crossref] [PubMed]
- Tang C, Lee WC, Reuben A, et al. Immune and Circulating Tumor DNA Profiling After Radiation Treatment for Oligometastatic Non-Small Cell Lung Cancer: Translational Correlatives from a Mature Randomized Phase II Trial. Int J Radiat Oncol Biol Phys 2020;106:349-57. [Crossref] [PubMed]
- Apte RN, Dotan S, Elkabets M, et al. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev 2006;25:387-408. [Crossref] [PubMed]
- De Miguel MP, Fuentes-Julián S, Blázquez-Martínez A, et al. Immunosuppressive properties of mesenchymal stem cells: advances and applications. Curr Mol Med 2012;12:574-91. [Crossref] [PubMed]
- Wang R, Zhang J, Chen S, et al. Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung Cancer 2011;74:188-96. [Crossref] [PubMed]
- Kinoshita T, Ishii G, Hiraoka N, et al. Forkhead box P3 regulatory T cells coexisting with cancer associated fibroblasts are correlated with a poor outcome in lung adenocarcinoma. Cancer Sci 2013;104:409-15. [Crossref] [PubMed]
- Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 2007;25:5287-312. [Crossref] [PubMed]
- Heller G, McCormack R, Kheoh T, et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J Clin Oncol 2018;36:572-80. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov 2016;6:479-91. [Crossref] [PubMed]
- Bayarri-Lara C, Ortega FG, Cueto Ladrón de Guevara A, et al. Circulating Tumor Cells Identify Early Recurrence in Patients with Non-Small Cell Lung Cancer Undergoing Radical Resection. PLoS One 2016;11:e0148659 [Crossref] [PubMed]
- Lin PP, Gires O, Wang DD, et al. Comprehensive in situ co-detection of aneuploid circulating endothelial and tumor cells. Sci Rep 2017;7:9789. [Crossref] [PubMed]
- Liu X, Li J, Cadilha BL, et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci Adv 2019;5:eaav4275 [Crossref] [PubMed]
- Hida K, Maishi N, Annan DA, et al. Contribution of Tumor Endothelial Cells in Cancer Progression. Int J Mol Sci 2018;19:1272. [Crossref] [PubMed]
- Lin PP. Aneuploid CTC and CEC. Diagnostics (Basel) 2018;8:26. [Crossref] [PubMed]
- Menter DG, Tucker SC, Kopetz S, et al. Platelets and cancer: a casual or causal relationship: revisited. Cancer Metastasis Rev 2014;33:231-69. [Crossref] [PubMed]
- Schumacher D, Strilic B, Sivaraj KK, et al. Platelet-derived nucleotides promote tumor-cell transendothelial migration and metastasis via P2Y2 receptor. Cancer Cell 2013;24:130-7. [Crossref] [PubMed]
- Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer 2011;11:123-34. [Crossref] [PubMed]
- Gareau AJ, Brien C, Gebremeskel S, et al. Ticagrelor inhibits platelet-tumor cell interactions and metastasis in human and murine breast cancer. Clin Exp Metastasis 2018;35:25-35. [Crossref] [PubMed]
- Yang L, Lv Z, Xia W, et al. The effect of aspirin on circulating tumor cells in metastatic colorectal and breast cancer patients: a phase II trial study. Clin Transl Oncol 2018;20:912-21. [Crossref] [PubMed]
- Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 2018;18:134-47. [Crossref] [PubMed]
- Tohme S, Yazdani HO, Al-Khafaji AB, et al. Neutrophil Extracellular Traps Promote the Development and Progression of Liver Metastases after Surgical Stress. Cancer Res 2016;76:1367-80. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Park S, Holmes-Tisch AJ, Cho EY, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol 2009;4:809-15. [Crossref] [PubMed]
- Shah RB, Mehra R, Chinnaiyan AM, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res 2004;64:9209-16. [Crossref] [PubMed]
- Drake JM, Graham NA, Lee JK, et al. Metastatic castration-resistant prostate cancer reveals intrapatient similarity and interpatient heterogeneity of therapeutic kinase targets. Proc Natl Acad Sci U S A 2013;110:E4762-9. [Crossref] [PubMed]
- Han HS, Eom DW, Kim JH, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer 2011;12:380-6. [Crossref] [PubMed]
- Quéré G, Descourt R, Robinet G, et al. Mutational status of synchronous and metachronous tumor samples in patients with metastatic non-small-cell lung cancer. BMC Cancer 2016;16:210. [Crossref] [PubMed]
- Vignot S, Frampton GM, Soria JC, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol 2013;31:2167-72. [Crossref] [PubMed]
- Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science 1977;197:893-5. [Crossref] [PubMed]
- Chambers AF, Hill RP, Ling V. Tumor heterogeneity and stability of the metastatic phenotype of mouse KHT sarcoma cells. Cancer Res 1981;41:1368-72. [PubMed]
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- Kratz JR, Haro GJ, Cook NR, et al. Incorporation of a Molecular Prognostic Classifier Improves Conventional Non-Small Cell Lung Cancer Staging. J Thorac Oncol 2019;14:1223-32. [Crossref] [PubMed]
- Lazzari C, Gregorc V, Karachaliou N, et al. Mechanisms of resistance to osimertinib. J Thorac Dis 2020;12:2851-8. [Crossref] [PubMed]
- Gan GN, Weickhardt AJ, Scheier B, et al. Stereotactic radiation therapy can safely and durably control sites of extra-central nervous system oligoprogressive disease in anaplastic lymphoma kinase-positive lung cancer patients receiving crizotinib. Int J Radiat Oncol Biol Phys 2014;88:892-8. [Crossref] [PubMed]
- Yu HA, Sima CS, Huang J, et al. Local therapy with continued EGFR tyrosine kinase inhibitor therapy as a treatment strategy in EGFR-mutant advanced lung cancers that have developed acquired resistance to EGFR tyrosine kinase inhibitors. J Thorac Oncol 2013;8:346-51. [Crossref] [PubMed]


