
Homologous recombination pathway gene variants identified by tumor-only sequencing assays in lung carcinoma patients
Highlight box
Key findings
• Higher variant allele fractions (VAF) for homologous recombination (HR) repair pathway gene variants identified in lung cancers, with tumor-only sequencing, may suggest germline origin.
What is known and what is new?
• HR gene variants can be identified in lung cancer, but their clinical significance is unclear.
• A subset of HR gene variants identified in lung cancer were confirmed to be of germline origin, which may be associated with familial cancer risks.
What is the implication, and what should change now?
• Genetic counseling and followup germline testing may be of value in a subset of patients for whom HR gene variants were identified with tumor-only sequencing, depending on the VAF and/or clinical/family history.
Introduction
The homologous recombination (HR) DNA repair pathway plays a key role in double-stranded DNA break repair. Briefly, this pathway senses double-stranded DNA breaks by the MRN (MRE11-RAD50-NBS1) complex, followed by ATM activation, extensive DNA processing (end resection, facilitated by BRCA1-PALB2-BRCA2 and RAD51 paralogs), homology search (after loading of RPA and RAD51), and subsequent homologous template-dependent repair (1-5). Somatic variants in the HR repair pathway genes, including BRCA1 and BRCA2 gene variants, have been identified in some non-small cell lung cancer (NSCLC) specimens, but the clinical significance of these variants remains poorly understood. By contrast, the HR pathway gene variants are of marked prognostic and predictive significance in several other cancers. Germline HR gene mutations are associated with increased cancer risk exemplified by the hereditary breast and ovarian cancer (HBOC) syndrome (6). However, many laboratories perform tumor-only sequencing, where the germline-somatic distinction can be difficult. Higher variant allele fractions in tumor (VAFs; which is the number of times a variant is detected relative to the overall number of times a genomic position is sequenced) may suggest germline origin, although confirmation through formal germline testing is always recommended (7).
Lung cancer is generally understood to be related predominantly related to environmental factors. Only a small number of germline variants have been associated with increased lung cancer risk (e.g., EGFR p.T790M) (8). While the American College of Medical Genetics and Genomics provides guidance on referral for cancer predisposition assessment (9), lung cancer is not addressed in the practice guideline. Lung cancer patients do not routinely undergo genetic counseling. In assessing family cancer history, significance of lung cancer cases in relatives is often unclear. In this study, we report HR pathway gene variants detected in tumors from 56 patients with NSCLC, and correlate these findings with clinical, pathological and molecular data. We present this article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-749/rc).
Methods
Case selection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was performed with the approval of University of Pennsylvania’s Ethics Board (Protocol No. 834224) and the requirement for patient consent was waived by the research ethics board, considering the retrospective nature of the study. We examined all cases sequenced at the Center for Personalized Diagnostics (CPD; University of Pennsylvania, from September 2016 to October 2019) and filtered for cases of interest using the algorithm outlined in Figure S1. Cases were selected based on the tissue assignment (by referring physician) and on the sequencing panel performed on the submitted specimen (see below). In total, 1,109 cases of “lung” cancers were sequenced using our solid tumor panel, which targets 152 genes (encompassing ~0.5 Mbp of genomic DNA). The resultant mutational data were plotted as waterfall and lollipop plots using R packages (GenVisR, version 3.11). Chart reviews were performed for the 56 patients with a confirmed diagnosis of lung cancer in whom we found variants in the HR repair pathway, and abstracted for details of pathology, any germline testing data, past medical history including cigarette smoking, and family cancer history. All data analyzed are available in Table S1.
Statistical analyses
Comparisons of continuous data between two groups were performed the Mann-Whitney test. Comparison of categorical data between two groups was performed using the Fisher’s exact test.
Massively parallel sequencing assays
Molecular workup for a new patient with lung cancer at the Hospital of the University of Pennsylvania has rapidly evolved over the past 10 years. The most recent version of the workup algorithm entails detection of single nucleotide variants and other small (i.e., less than approximately 30 bp) genetic alterations, using a 152-gene sequencing panel (solid panel version 2, Table S2). Fusion transcripts, including ALK, RET, ROS1, and NTRK fusions, are detected using a 55 gene-target fusion panel (Table S3), and/or fluorescence in situ hybridization. MET exon 14 skipping mutations (i.e., exons joined out of order or point mutations known to cause exon skipping events) are detected by these panels. Genetic variants detected are internally classified as disease-associated, probably disease-associated, variant of unknown significance (VUS), likely benign, or benign, and reported as either disease associated or VUS (which encompasses probably disease-associated, VUS, and likely benign). While the concepts are not formally equivalent, this classification scheme is similar to the Association for Molecular Pathology (AMP)/American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) classification system (10); disease-associated and probably disease-associated variants would be largely equivalent to AMP/American College of Medical Genetics (ACMG) tier I–II variants (i.e., therapeutic, prognostic or diagnostic significance). For example, strong clinical significance for BRCA1/2 gene variants may be related to variants’ known association with familial cancer risks, or response to PARP inhibitors, among other lines of evidence. Other variants would be classified as tier III (VUS) or IV (likely benign and benign); these latter variants were excluded from further analysis. Interpretation and tiering of variants was performed in the context of available data, including which include gene-specific (i.e., BRCA1/2) databases. Of note, the next generation sequencing (NGS) workflow is a tumor-only sequencing assay, and the distinction between variants of germline and somatic derivation is not made. Tumor mutational burden (TMB) was assessed using an internally developed algorithm (manuscript in preparation).
Results
Data review and case selection
The sequencing panel includes 14 genes in the HR repair pathway, namely ATM, ATRX, BRCA1, BRCA2, BRIP1, CHEK2, MRE11A, NBN, PALB2, RAD50, RAD51, RAD51B, RAD51C, and RAD51D, ranging from genes with well-established roles in HR repair (e.g., BRCA1/2) to genes with more recently recognized roles [e.g., ATRX (11)]. Following retrospective review of molecular profiling data for 1,109 cases of “lung cancers”, we identified 61 disease-associated/probably disease-associated variants in 56 unique patients with NSCLC. ATM gene variants were most common (24/61 variants), followed by BRCA1 (9 variants), ATRX (4 variants) and BRCA2 (4 variants) (Figure 1). Multiple HR gene pathway variants were identified in five patients, three of whom had two disease-associated variants in the ATM gene. The 56 unique cases comprised 49 adenocarcinomas, one squamous cell carcinoma (SCC), and six carcinomas not otherwise specified (NOS). The patient ages ranged from 39 to 91 years (median 68) at diagnosis. Cigarette smoking history was available in 55 patients, of which 50 patients had a positive smoking history, with 5 never-smokers. More detailed smoking history was available for 41 patients, among which the average exposure was 39.5 pack-years. Among oncogenic driver variants in our 56 patients with HR repair pathway mutations, co-mutations in KRAS were the most common (30/56 patients), followed by EGFR and MET (2 patients each). Single cases of ALK, NRAS and ERBB2-driven cases were also identified. No driver genetic mutations were identified in 19 cases. TMB ranged from 0 to 34.3 mutations/megabase (average 7.4).
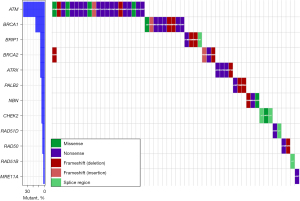
Variants with higher allele fractions
We filtered the variants based on VAFs—we reasoned that HR gene variants seen at higher VAFs are more likely to be of germline origin. Seventeen DNA variants from 17 patients were observed at VAFs ≥30%. While germline variants are often identified at VAF of approximately 50%, VAF of 30% was chosen to increase the sensitivity for identifying potentially germline variants, as VAFs for germline variants can range widely, related to both biologic (e.g., copy number changes) and technical (e.g., low read depth) issues. Of the 17 patients with HR pathways gene variants at ≥30% VAF, four patients had a prior personal cancer history (Figure 2). Family history was available for 16/17 high VAF patients, with family history information limited in one patient. By the National Comprehensive Cancer Network (NCCN) HBOC criteria (12), family history would be considered significant in two patients. That is, the patient would have met the criteria for germline testing; one such patient had a history of ovarian cancer in a 1st-degree relative, and another patient had a 2nd-degree relative with ovarian cancer. Breast cancer family history was positive in seven patients, 6/7 cases of which were in 1st-degree relatives, although the ages at diagnosis and cancer subtype (e.g., triple-negative) were not available. Similarly, prostate cancer family history was positive for three patients, but information regarding the specific disease attributes (Gleason grade, disease burden, clinical course) was unavailable. Family history of pancreatic cancer was positive in one patient (2nd-degree relative). In addition to the typical HBOC-associated cancers, family history of lung cancer was identified in 4/16 patients, all such patients having at least one affected 1st-degree family member.
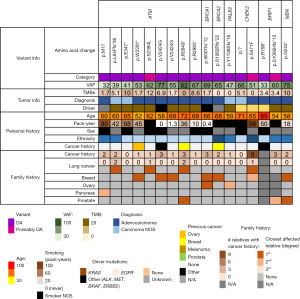
Of the 14 HR pathway genes examined, only variants in seven genes were observed at a VAF ≥30%, with ATM gene variants being most common (9/17 patients). In the germline setting, ATM is generally considered an intermediate cancer risk gene, with the exception of ATM p.V2424G (NM_000051.4: c.7271T>G) that is associated with higher cancer risk when compared to other ATM variants (13). ATM p.V2424G was observed in two patients at VAFs of 55% and 77%; both patients had a history of breast cancer in their families. Seven other ATM disease-associated/probably disease-associated variants were identified, comprised largely of disruptive frameshift and nonsense variants. One CHEK2 variant (NM_001005735.1: c.1412C>T; p.S471F, equivalent to NM_007194.4: c.1283C>T) stood out from the other variants, occurring in a patient from a family with nine members with a cancer history.
High VAF HR gene variants and correlations with germline/repeat somatic sequencing results
Three patients underwent germline testing, which was initiated based on the somatic sequencing results. All three patients were positive for pathogenic germline variants (Table 1). Among the 14 HR genes included in our study, the highest cancer risks are associated with BRCA1, BRCA2, and PALB2 (14-16), and a germline pathogenic variant was identified in each of the high-risk genes. Examining the somatic sequencing results, BRCA1 and BRCA2 variants were observed at VAFs of 69% and 65%, respectively, suggesting that loss-of-heterozygosity (LOH) may have occurred in the tumors. In two patients, lung cancer was their first (sentinel) cancer, with no personal history of other cancers. One BRCA1 disease-associated variant (NM_007300.4: c.1799delT; p.I600Tfs*12) was confirmed to be germline in a patient who presented at age 66 with lung cancer; the family cancer history was notable for ovarian and breast cancer in 1st degree relatives (mother and sister, respectively). In a patient with a germline BRCA2 variant (NM_000059.4: c.5946delT; p.S1982Rfs*22), the past medical history was significant for a previous triple-negative breast carcinoma that preceded the lung cancer by 12 years. A PALB2 variant (NM_024675.4: c.3323delA; p.Y1108Sfs*16) was confirmed to be germline in a patient who presented with lung cancer at age 59; family cancer history was notable for ovarian cancer in her maternal grandmother. All three patients had a cigarette smoking history, with the pack-years history unknown in two patients.
Table 1
| Age/sex (smoking history) | Germline genetics (annotation) | Clinical history |
|---|---|---|
| 66M (0.4 PY) | BRCA1 NM_007300.4: c.1799delT (p.I600fs*12, pathogenic) | Personal: none |
| Family: ovarian cancer (mother@47), breast (sister@62), mesothelioma (father@84) | ||
| 68F (“Former”, PY NOS) | BRCA2 NM_000059.4: c.5946delT (p.S1982Rfs*22, pathogenic) | Personal: breast carcinoma (56 years, triple negative), ovarian fibroma (47 years) |
| Family: breast (mother), uterus (mother) | ||
| 59F (“Former”, PY NOS) | PALB2 NM_024675.4: c.3323delA (p.Tyr1108Serfs*16, pathogenic) | Personal: none |
| Family: ovarian cancer (maternal grandmother) |
#, germline variant terminology differs from somatic variant classification terminology. However, in this context, “pathogenic” germline variant can be considered being equivalent to “disease-associated”. @: diagnosed at age. M, male; PY, pack-year; F, female; NOS, not otherwise specified.
Three other patients had undergone repeat somatic (tumor-only) sequencing without germline testing (Table 2). If HR pathway gene variants are detected in two metachronous tumors from the same patient, this increases the likelihood that the variants are of germline origin. For all three patients who had undergone repeat somatic sequencing, the HR pathway gene variants were consistently detected. Two of these patients had the ATM p.V2424G variant, and one patient had a BRIP gene variant. In one patient, the ATM p.V2424G variant was identified in two definitively metachronous, independent tumors that harbored two different KRAS mutations, lending further support to the ATM gene variant being germline in the patient.
Table 2
| Variant | Detected on repeat assay? | VAF | Time between samples | Shared driver gene? | |
|---|---|---|---|---|---|
| Initial | Repeat | ||||
| ATM NM_000051.4: c.7271T>G (p.V2424G) | Yes | 77% | 78% | 3 years | N/A* |
| ATM NM_000051.4: c.7271T>G (p.V2424G) | Yes | 55% | 56% | 1 year | No (KRAS)** |
| BRIP1 NM_032043.2: c.3196delT (p.S1066Hfs*12) | Yes | 60% | 80% | 1 year | Yes (KRAS) |
#, the table compares data from metachronous, potentially independent (vs. recurrent) tumors that underwent molecular profiling from three different patients; *, N/A: not applicable—no driver genetic event identified in either samples; **, two different KRAS driver mutations (c.35G>A; p.G12D and c.34G>T; p.G12C), consistent with two independent tumors. HR, homologous recombination; VAF, variant allele fraction.
Comparison of patients with high VAF vs. low VAF tumors
The above data suggested at least six of our patients (who had undergone germline or repeat somatic sequencing) likely harbor germline variants in HR pathway genes, confirming the potential utility of a VAF ≥30% as a screen to identify potential germline variants. By contrast, the HR pathway genes observed at low VAFs are more likely to be truncal, possibly passenger mutations, acquired during tumor progression/evolution. Given that this distinction may be helpful in patient selection for germline testing, we compared the patients with high vs. low VAF variants for clinicopathological features that may further aid in the distinction. When we compared the patients with HR gene variants identified at low or high VAFs, the two groups did not differ with respect to age at diagnosis, proportion of 20+ pack-year cigarette smokers, or mean pack-years (where the information was available, Table 3). Also, their tumors did not differ significantly with respect to proportion with/without targetable genetic mutations; KRAS was the most common oncogenic driver mutation in both groups. Interestingly, the TMBs were significantly lower in the VAF ≥30% group (Mann-Whitney P=0.0036), although the ranges were wide for both groups (0 to 12.0/Mbp for VAF ≥30%, and 0 to 34.3/Mbp for VAF <30%). Therefore, none of the conventional clinical, pathological or molecular attributes appeared to be a useful flag for potential germline variants in HR pathway genes, with the exception of differences in VAFs and TMBs.
Table 3
| Groups features | VAF <30% | VAF ≥30% | P values (test) |
|---|---|---|---|
| Number | 39 patients | 17 patients | |
| Age at diagnosis | Mean age 67.8 years (SD 10.7) | Mean 64.5 years (SD 8.9) | 0.1031 (Mann-Whitney) |
| Cigarette smoking history | 34/38 (89.5%) positive history | 16/17 (94.1%) positive | 1.000 (Fisher’s exact test) |
| 38.7 mean pack-years (SD 21.1, n=30)** | 41.7 mean pack-years (SD 28.4, n=11)** | 0.9283 (Mann-Whitney) | |
| 26/30 (86.7%) with ≥20 pack year smoking history | 7/11 (63.6%) with ≥20 pack year smoking history | 0.1777 (Fisher’s exact test) | |
| Driver | |||
| Non-targetable* | 20 KRAS, 1 NRAS, 7 none, 7 unknown | 10 KRAS, 3 none, 2 unknown | 1.000 (Fisher’s exact test) |
| Targetable | 2 EGFR, 2 MET | 1 ALK, 1 ERBB2 | |
| TMBs (mutations per megabase, /Mbp), mean ± SD | 8.8±6.2 | 4.2±6.6 | 0.0036 (Mann Whitney) |
Note: KRAS variants include the KRAS p.G12C variant, which was not a targetable gene variant at the time of patient work-up. *, generally associated with little or no smoking history; **, n indicates the number of patients with known pack-years. VAF, variant allele fraction; HR, homologous recombination; SD, standard deviation; TMB, tumor mutational burden.
Discussion
We report HR pathway gene variants (categorized as disease-associated or probably disease-associated at our institution) in 56/1,109 (5.0%) lung cancer specimens, with the gene variants being observed over a wide range of VAFs. Most variants (44/61, 72.1%) were observed at VAF <30%; such variants are likely passenger mutations acquired with tumor evolution, unlikely to be of germline origin, and unlikely to be associated with LOH. The remaining 17 patients’ tumors harbored HR gene variants at VAFs ≥30%, where the higher VAFs suggested that some of the variants may be germline in origin. Indeed, a subset of these variants, which included disease-associated BRCA1/2, or PALB2 mutations, as well as ATM variants, were associated with even higher VAFs and often with significant personal and/or family cancer history. While the VAF cut-off of 30% was useful in our study, its sensitivity and specificity in use for flagging potentially germline variants are to be studied further.
Genetic counseling for lung cancer patients can be challenging for numerous reasons, including the generally short survival span for these patients; the five-year survival for lung cancer (~21%) differs drastically from breast cancer (90%) [Surveillance, Epidemiology, and End Results (SEER) data, accessed June 2020]. Discussion of “familial lung cancer” is also limited in the literature, with the EGFR p.T790M variant as the sole well-established risk-elevating allele (8). Many lung cancer patients also have a cigarette smoking history, which also may be a shared risk factor amongst family members through first and/or second-hand smoking. Our data demonstrate that lung cancer patients can harbor pathogenic, germline HR pathway gene variants and that somatic testing can point to their existence. Our data suggest that patient characteristics such as older age and cigarette smoking history are unlikely to be helpful in identifying germline HR pathway variants. As such, germline testing should be considered in all patients in whom pathogenic high-VAF variants are identified by somatic testing. Although only three patients with HR pathway gene variants at high VAF underwent germline testing, it is noteworthy that all three were positive for HR gene variants, suggesting that lung cancer can be the sentinel cancer in a patient with a pathogenic BRCA1 or PALB2 gene variant.
Among the 14 HR genes analyzed in our study, ATM variants were the most common, which may be related to the large size of the ATM gene (146,619 bp, based on hg19). However, a few of the other HR pathway genes are comparable in size, including BRCA1 (125,951 bp). Many of the observed ATM gene variants in our patients with lung cancer are highly suspicious for germline origin, based on the high VAFs and clinical histories. The repeat somatic sequencing data are particularly suggestive, especially in the patient with two metachronous, independent tumors harboring the same ATM gene variant. Several ATM single-nucleotide polymorphisms have been linked to increased lung cancer risk, including rs189037 (c.-111G>A on the transcript NM_000051.3), rs664677 (c.3078-77C>T) and rs664143 (c.8850+60A>G) (17,18). More recently, a case-control association study examining 1,083 lung adenocarcinoma patients identified rare, deleterious, germline ATM variants more frequently in patients with lung adenocarcinoma (odds ratio of 4.6) (19). The ATM gene variants identified in that study did not overlap with the variants identified in our study, partly related to our exon-centric sequencing panel design. Our data also support the concept that ATM gene variants may be related to increased familial lung cancer risk, likely further exacerbated by cigarette smoking.
Conclusions
In summary, we demonstrate that pathogenic HR pathway gene variants are identified in a subset of patients with primary lung cancer undergoing routine tumor-only sequencing as part of standard of care. Among the patients in whom these HR pathway variants are observed at higher VAFs, including patients with ATM gene variants, further germline testing may be of value to the patients and their relatives, regardless of patient’s age at diagnosis, and cigarette smoking history.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-749/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-749/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-749/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-749/coif). JJR reports holding stock/stock options in Pfizer and Eli Lilly personally and serves as a member representative on an external advisory stakeholder for a grant entitled “Randomized trial of universal vs. guideline-directed germline testing among young adults with cancer” which is an NCI Cancer Moonshot Approaches to Identify and Care for Individuals with Inherited Cancer Syndromes (U01). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was performed with the approval of University of Pennsylvania’s Ethics Board (Protocol No. 834224) and the requirement for patient consent was waived by the research ethics board, considering the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 2013;5:a012716. [Crossref] [PubMed]
- Densham RM, Garvin AJ, Stone HR, et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat Struct Mol Biol 2016;23:647-55. [Crossref] [PubMed]
- Isono M, Niimi A, Oike T, et al. BRCA1 Directs the Repair Pathway to Homologous Recombination by Promoting 53BP1 Dephosphorylation. Cell Rep 2017;18:520-32. [Crossref] [PubMed]
- Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochim Biophys Acta 2014;1846:263-75. [PubMed]
- Harris JL, Rabellino A, Khanna KK. RAD51 paralogs promote genomic integrity and chemoresistance in cancer by facilitating homologous recombination. Ann Transl Med 2018;6:S122. [Crossref] [PubMed]
- Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In: Adam MP, Mirzaa GM, Pagon RA, et al. editors. GeneReviews®. Seattle, WA, USA: University of Washington, 1998.
- Li MM, Chao E, Esplin ED, et al. Points to consider for reporting of germline variation in patients undergoing tumor testing: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2020;22:1142-8. [Crossref] [PubMed]
- Kanwal M, Ding XJ, Cao Y. Familial risk for lung cancer. Oncol Lett 2017;13:535-42. [Crossref] [PubMed]
- Hampel H, Bennett RL, Buchanan A, et al. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med 2015;17:70-87. [Crossref] [PubMed]
- Li MM, Datto M, Duncavage EJ, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4-23. [Crossref] [PubMed]
- Juhász S, Elbakry A, Mathes A, et al. ATRX Promotes DNA Repair Synthesis and Sister Chromatid Exchange during Homologous Recombination. Mol Cell 2018;71:11-24.e7. [Crossref] [PubMed]
- NCCN. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic. NCCN Clinical Practice Guidelines in Oncology 2023.
- Jerzak KJ, Mancuso T, Eisen A. Ataxia-telangiectasia gene (ATM) mutation heterozygosity in breast cancer: a narrative review. Curr Oncol 2018;25:e176-80. [Crossref] [PubMed]
- Easton DF, Pharoah PD, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015;372:2243-57. [Crossref] [PubMed]
- Macedo GS, Alemar B, Ashton-Prolla P. Reviewing the characteristics of BRCA and PALB2-related cancers in the precision medicine era. Genet Mol Biol 2019;42:215-31. [Crossref] [PubMed]
- Rebbeck TR, Mitra N, Wan F, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 2015;313:1347-61. [Crossref] [PubMed]
- Yan Z, Tong X, Ma Y, et al. Association between ATM gene polymorphisms, lung cancer susceptibility and radiation-induced pneumonitis: a meta-analysis. BMC Pulm Med 2017;17:205. [Crossref] [PubMed]
- Zhao ZL, Xia L, Zhao C, et al. ATM rs189037 (G > A) polymorphism increased the risk of cancer: an updated meta-analysis. BMC Med Genet 2019;20:28. [Crossref] [PubMed]
- Esai Selvan M, Zauderer MG, Rudin CM, et al. Inherited Rare, Deleterious Variants in ATM Increase Lung Adenocarcinoma Risk. J Thorac Oncol 2020;15:1871-9. [Crossref] [PubMed]


