
High NHLRC2 expression is associated with shortened survival in lung adenocarcinoma
Highlight box
Key findings
• Immunohistochemical NHL repeat (named after NCL-1, HT2A and LIN-41)-containing protein 2 (NHLRC2) expression was higher in lung adenocarcinoma compared to squamous cell carcinoma.
• High NHLRC2 expression was associated with poor survival in lung adenocarcinoma patients.
What is known and what is new?
• Certain variants of NHLRC2 have been associated with a multiorgan disease with severe lung fibrosis in early childhood.
• In this study, the expression pattern of NHLRC2 in lung tissue samples from lung adenocarcinoma and squamous cell carcinoma patients was described for the first time.
• The NHLRC2 expression was compared with the clinical and histological data of the lung cancer patients.
What is the implication, and what should change now?
• NHLRC2 may function as a prognostic biomarker for lung adenocarcinoma patients.
Introduction
Despite the advances in cancer therapy, lung cancer is the leading cause of cancer deaths worldwide accounting 18% of the total cancer deaths (1). The most common histological types of lung cancer are adenocarcinoma (ADC) and squamous cell carcinoma (SCC), and they are further divided into several subtypes (2). Although both ADC and SCC are classified as non-small cell lung cancer, they originate from different cell types and have several differences in biological patterns, molecular characteristics, genetic alterations, and therapeutic strategies (3,4). SCC mostly develops in smokers, while ADC is the most common type of lung cancer seen in nonsmokers.
NHL repeat (named after NCL-1, HT2A and LIN-41)-containing protein 2 (NHLRC2) is a 79 kDa protein consisting of a N-terminal thioredoxin (Trx)-like domain and a C-terminal NHL-repeat domain. Certain variants of NHLRC2 have been linked to a multiorgan disease with severe lung fibrosis in early childhood (OMIM #618278) (5-7). A loss of essential NHLRC2 has been shown to lead to failed gastrulation, amniotic folding, and embryonic lethality in mice (8). We have observed previously that the expression of NHLRC2 was increased in lung tissues of patients with idiopathic pulmonary fibrosis (IPF) (9). NHLRC2 has been identified as a differentially expressed gene in lung tissue samples between fast and slowly progressing IPF (10). High mRNA expression of NHLRC2 has been associated with better prognosis in lung ADC patients when combined with the expression of two other protein coding genes and one long non-coding RNA in homogenized lung tissue samples (11). The data generated from our previous microarray analysis revealed that NHLRC2 was expressed in stromal cells derived from ADC (Gene expression omnibus accession number GSE144338) (12). To our knowledge, however, expression pattern of NHLRC2 in lung cancer tissues has not been previously published.
The aim of this study was to examine and compare the expression patterns of NHLRC2 protein and mRNA in lung tissues of lung ADC or SCC patients by immunohistochemistry and mRNA in situ hybridization. Immunohistochemical NHLRC2 expression assessed by digital pathology image analysis software was compared with the patients’ clinical and histological data. Additionally, the percentage of NHLRC2-positive cancer cells was evaluated by semiquantitative analysis. NHLRC2 protein levels in lung cell lines and tissue homogenates were also measured by Western blot analysis. We present this article in accordance with the RECORD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-815/rc).
Methods
Patients
Lung tissue samples were retrieved from the files of the Biobank Borealis, and the Department of Pathology, Oulu University Hospital. A retrospective analysis was conducted on 111 SCC and 102 ADC patients who had surgical lung resection in the Oulu University Hospital between 1998 and 2007. The details of ADC patients have been presented previously (13). Clinical information, including age, sex, smoking history, pulmonary function test results, and follow-up data was gathered systematically from the medical records as described previously (13). A non-smoker was defined as a person who had smoked less than 100 cigarettes in his or her lifetime. Overall survival (OS) was defined as the time from the date of diagnosis to the date of death from any cause or last follow-up and disease specific survival (DSS) as the time from the date of diagnosis to the date of death from lung cancer. Additionally, control samples were derived from ten non-smoking ADC patients from histologically normal-looking peripheral lung.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). A favorable statement of the study protocol was given by the Ethical Committee of Northern Ostrobothnia Hospital District in Oulu (2/2008, amendments 12/2014, 2/2015, 2/2018 and 6/2022). National Supervisory Authority for Welfare and Health has approved the research use of paraffin-embedded tissue samples (Dnro: V/25090/2019) and individual consent for this retrospective analysis was waived. For collection of cell culture materials, all subjects gave their written informed consent.
Histopathology
Each tumor was re-evaluated according to the 2015 World Health Organization classification based on alcian blue mucin and hematoxylin and eosin stains (14). The details of predominant growth pattern analysis of ADC have been presented previously (13,15). Additionally, the following histological parameters were analyzed: desmoplasia, nuclear atypia, tumor necrosis, mitotic activity, and lymphovascular invasion as described previously (15). The pathological stage was redetermined according to the TNM (tumor, node, metastasis) classification of malignant tumors (Union for International Cancer Control/American Joint Committee on Cancer, 7th edition) as described previously (13,16,17).
Immunohistochemistry
Formalin-fixed and paraffin-embedded lung tissue specimens were cut into 3.5-µm thick sections, which were de-paraffinized in xylene and rehydrated in decreasing concentrations of ethanol. Following heat-induced or enzymatic antigen retrieval, sections were stained by using Dako EnVision Flex Kit (Dako, Glostrup, Denmark) with diaminobenzidine (DAB+) chromogen. Antibodies are listed in Table S1. Sections were counterstained with Mayer’s hematoxylin (Sigma-Aldrich, Steinheim, Germany). For negative controls, primary antibodies were replaced with a rabbit isotype control (Invitrogen, Carlsbad, USA). The expression of NHLRC2 was compared to the expression of collagen α1(IV) chain on the basis of the results of our previous study on the microarray analysis of lung stromal cells (12), and similarly to our previous study on IPF (9). Cluster of differentiation 68 (CD68), and alpha-smooth muscle actin (α-SMA) antibodies were used to identify macrophages and myofibroblasts, respectively.
Whole slide images were acquired at 40× magnification with a NanoZoom S60 scanner (Hamamatsu, Hamamatsu city, Japan) by Transgenic and Tissue Phenotyping core facility, Biocenter Oulu, University of Oulu.
Digital image analysis of immunohistochemical NHLRC2 expression
The area of NHLRC2-positive staining (weak, moderate, and strong) in all cell types within cancer tissue in relation to the total cancer tissue area was determined by using Visiopharm digital pathology image analysis software (Visiopharm Integrator System, Hoersholm, Denmark) provided by Transgenic and Tissue Phenotyping core facility, Biocenter Oulu, University of Oulu. Areas containing necrosis were excluded from analysis. In controls, the NHLRC2-positive area was determined in relation to the total tissue section area as described previously (9).
Scoring of immunostaining of NHLRC2
To further study the NHLRC2 expression specifically in cancer cells, digitized lung tissue specimens were examined by using NDP.view2 (Hamamatsu). The percentage of NHLRC2-positive cancer cells was scored as follows: negative, less than 25% of cells positive, 25–49% of cells positive, 50–75% of cells positive, and over 75% of cells positive. The extent of NHLRC2 staining in tumor cells in ADCs was compared to that in SCCs.
mRNA in situ hybridization
RNAscope 2.5 HD assay—RED and probe Hs-NHLRC2 (555721, Advanced cell diagnostics, Newark, CA, USA) were used for detection of NHLRC2 mRNA from 4-µm thick formalin-fixed and paraffin-embedded lung tissue sections derived from ADC (n=4) and SCC (n=3) patients according to the manufacturer’s instructions as described previously (9,18). Probe for the bacterial gene 4-hydroxy-tetrahydrodipicolinate reductase (dapB, 310043, advanced cell diagnostics) was used as negative control to assess background signals. Probe for the endogenous housekeeping gene ubiquitin C (UBC, 310041, Advanced cell diagnostics) was used as positive control to assess assay procedure. Whole slide images were acquired as described in section Immunohistochemistry above and examined by using NDP.view2.
Cell culture
The expression levels of NHLRC2 in primary stromal and epithelial cell lines were compared in vitro. Stromal cells from lung cancer patients from tumor (ADC n=2, SCC n=2) and corresponding tumor free peripheral lung (ADC n=2, SCC n=2) were cultured as described previously (19,20). The cells were cultured in Minimum essential medium Eagle (α modification) (Sigma-Aldrich) supplemented with 2 mM L-glutamine, 10 mM HEPES, 100 U/mL penicillin, 0.1 g/L streptomycin, 2.5 mg/L amphotericin B (all from Sigma-Aldrich), and 13% heat-inactivated fetal bovine serum (FBS-Good, Pan Biotech, Aidenbach, Germany) at 37 ℃ in humidified atmosphere containing 5% CO2. According to our electron microscopic analyses published previously, these cell lines are mixtures of fibroblasts and myofibroblasts (19, 20). For experiments, stromal cells were plated at a density of 3,300 cells/cm2 and cultured for four days at passages five to six.
Normal human primary bronchial/tracheal epithelial cells (PBTE) [American Type Culture Collection (ATCC), Virginia, USA, PCS-300-010] and normal human primary small airway epithelial cells (SAEC, ATCC, PCS-301-010) were cultured in airway cell basal medium supplemented with bronchial epithelial growth kit (ATCC). PBTE were used for experiments at passage five and SAEC at passage seven. Epithelial lung cancer cell lines H1650 [ATCC CRL-5883, Research Resource Identifier (RRID):CVCL_1483, minimally invasive lung ADC], SK-LU-1 (ATCC HTB-57, RRID:CVCL_0629, ADC) and SK-MES-1 (ATCC HTB-58, RRID:CVCL_0630, SCC) were cultured in Minimum essential medium Eagle (α modification) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 10 mM HEPES, 100 U/mL penicillin, 0.1 g/L streptomycin, and 2.5 mg/L amphotericin B.
Immunoblotting
NHLRC2 protein levels in lung tissue samples collected from ADC (n=3) and SCC (n=2) patients from tumor and corresponding tumor free peripheral lung as well as lung cell lines were studied by immunoblotting. Frozen lung tissue samples were homogenized in 1.5% dodecyl maltoside (Thermo Fisher Scientific, Vilnius, Lithuania, in phosphate buffered saline) supplemented with a protease inhibitor cocktail tablet (Roche, Mannheim, Germany) by sonication and centrifuged at 20,000 g for 20 min. Cell lysates were prepared as described previously by using 1.5% dodecyl maltoside (9). Supernatants were collected and protein concentrations of tissue homogenates and cell lysates were determined by DC Protein Assay Kit (Bio-Rad Laboratories, Inc., USA) in accordance with the manufacturer’s guidelines. Twenty µg of protein per sample were loaded with Bolt LDS sample buffer (Thermo Fisher Scientific) onto a polyacrylamide gel (Invitrogen Bolt Bis-Tris Mini Protein Gels, Thermo Fisher Scientific). After electrophoresis, the proteins were transferred onto nitrocellulose membrane (0.45 µm, Optitran reinforced NC, Whatman Schleicher and Schuell, Dassel, Germany). Membranes were stained with TotalStain Q (NC, Azure Biosystems, Dublin, CA, USA). After blocking with 5% non-fat dry milk, the blots were incubated with primary antibody for NHLRC2 followed by labelled secondary antibody incubation (Table S1). Protein bands were visualized with an Azure 600 gel & blot imager (Azure Biosystems) and quantified using Image Studio Lite (LI-COR Biosciences). NHLRC2 expression levels were adjusted with the total protein stain.
Statistical analyses
Statistical analyses were performed with IBM SPSS Statistics for Windows, Version 28.0 (IBM Corp, Armonk, NY) and graphs were prepared with OriginPro, Version 2022 (OriginLab Corporation, Northampton, MA, USA). The data were presented as the means with standard deviation (SD) for those with a normal distribution, or as median values with 25% and 75% quartiles (interquartile range, IQR) for skewed variables. Independent samples t-test was used for normally distributed variables, Mann-Whitney U test for skewed variables, Wilcoxon signed ranks test for comparison of paired control and tumor tissues, and Fisher-Freeman-Halton test for comparison of categorial variables. Survival was analyzed using the Kaplan-Meier method, and log-rank test was used to evaluate differences in survival curves. Median values of NHLRC2 expression in each group were used as cut off values for Kaplan-Meier analysis. A P value less than 0.05 was considered statistically significant.
Results
Patients’ characteristics
The characteristics of the lung cancer patients included in the immunohistochemical analysis are presented in Table 1. Sixty-seven (66%) out of the 102 ADC patients and 97 (87%) out of the 111 SCC patients were men. Eighty-one (83%) out of 98 ADC patients and 107 (99%) out of 108 SCC patients with known smoking history were ex- or current smokers. The median follow-up time was 32.5 months (range, 0–172 months) in ADC patients and 46.0 months (range, 1–178 months) in SCC patients.
Table 1
| Parameters | Adenocarcinoma (n=102) | Squamous cell carcinoma (n=111) |
|---|---|---|
| Age (years), mean (SD) | 65 (8.70) | 67.86 (7.10) |
| Gender, n (%) | ||
| Male | 67 (65.69) | 97 (87.39) |
| Female | 35 (34.31) | 14 (12.61) |
| Smoking status, n (%)† | ||
| Non-smoker | 17 (16.67) | 1 (0.90) |
| Ex-smoker | 20 (19.61) | 56 (50.45) |
| Current smoker | 61 (59.80) | 51 (45.95) |
| Pack-years of ex- and current smokers, median (IQR)‡ | 30.00 (16.50–40.00) | 40.00 (26.00–50.00) |
| Stage, n (%)§ | ||
| IA | 30 (29.4) | 29 (26.1) |
| IB | 25 (24.5) | 17 (15.3) |
| IIA | 18 (17.6) | 30 (27.0) |
| IIB | 12 (11.8) | 7 (6.3) |
| IIIA | 14 (13.7) | 8 (7.2) |
| IV | 3 (2.9) | 2 (1.8) |
†, Information missing from 4 adenocarcinoma and 3 squamous cell carcinoma patients. ‡, Information missing from 39 adenocarcinoma and 26 squamous cell carcinoma patients. §, Information missing from 18 squamous cell carcinoma patients. IQR, interquartile range; n, number; SD, standard deviation.
NHLRC2 protein and mRNA expression patterns in lung cancer
The expression pattern of NHLRC2 was studied by immunohistochemistry and mRNA in situ hybridization. Negative to moderate cytoplasmic immunohistochemical NHLRC2 expression was detected in cancer cells in ADCs (n=102) and SCCs (n=111) (Figures 1-3). Moderate to strong cytoplasmic NHLRC2 expression was detected in inflammatory cells within cancer stroma. Macrophages within tumor were mainly weakly positive. Very weak NHLRC2 expression was occasionally detected in spindle shaped stromal cells. Endothelium was mainly negative. Normal and metaplastic epithelial cells of bronchi outside tumor were mainly positive for NHLRC2. The collagen α1(IV) expression pattern differed from that NHLRC2 since it was mainly detected extracellularly within the tumor stroma, while mainly weak expression was occasionally observed in cancer cells (Figure S1).
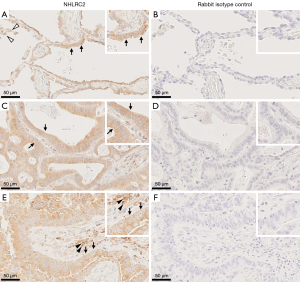


In control lung, moderate to quite strong NHLRC2 immunoreactivity was observed in alveolar macrophages, type II pneumocytes, and small airway epithelial cells. Endothelium and smooth muscle cells were mainly negative or weakly positive for NHLRC2.
The mRNA expression of NHLRC2 in tumor samples (n=7) was in line with the immunohistochemical expression. NHLRC2 expression was detected in some cancer cells and occasionally in some stromal cells within tumor (Figures S2,S3).
Association of immunohistochemical NHLRC2 expression with histological features
NHLRC2 expression in all cell types within tumor in relation to the total tumor area determined by image analysis software was higher in ADC (n=102, mean 17.72, SD 8.99) than in SCC (n=111, mean 12.04, SD 7.37, P<0.001) (Figure 4A). NHLRC2 expression was higher in ADC (mean 17.72, SD 8.99) and SCC (n=111, mean 12.04, SD 7.37) than in control (mean 4.23, SD 1.14, P<0.001 and P<0.001, respectively) (Figure 4A).
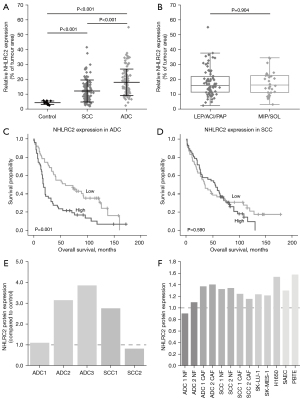
High NHLRC2 expression was associated with high mitotic activity in ADC (P=0.042) (Table S2). NHLRC2 expression did not correlate with other histopathological tumor parameters (nuclear atypia, tumor necrosis, desmoplasia, lymphovascular invasion) in ADC or SCC (Table S2).
The most frequent histologic ADC subtype was acinar (n=52, 51.0%), including seven tumors with cribriform pattern. Twenty-tree (22.5%) out of 102 ADCs were solid, eight (7.8%) were papillary, seven (6.9%) were micropapillary and one tumor (1.0%) was lepidic predominant ADC. Eleven (10.8%) out of 102 ADCs were invasive mucinous variants. Due to low number of cases in each group of ADC subtypes, they were divided into two groups based on the previous publication showing that micropapillary and solid ADCs had worse DSS than other histologic subtypes (13). The relative NHLRC2 expression did not differ between the histologic subtypes of ADC (Figure 4B). Majority of the SCC tumors were non-keratinizing type (n=99, 89.2%), while eight (7.2%) were keratinizing and four (3.6%) were basaloid type. NHLRC2 expression did not differ between SCC subtypes (median 11.28, IQR, 7.44–16.79; median 13.20, IQR, 8.85–20.08; median 9.30, IQR, 8.14–16.35, P=0.632).
Association of immunohistochemical NHLRC2 expression with clinical data
ADC patients having tumors with high relative NHLRC2 expression (over 16% of tumor area) had lower DSS and OS rate than patients having tumors with low NHLRC2 expression (less than 16% of tumor area, P=0.002 and P=0.001, respectively) (Table 2, Figure 4C). NHLRC2 expression did not correlate with survival in SCC (Table 2, Figure 4D). NHLRC2 expression did not correlate with stage, gender, smoking status, chronic obstructive pulmonary disease (COPD), age, or pulmonary function test results (Table S3).
Table 2
| Lung cancer type | NHLRC2 expression | 5-year survival (%) | P value | Mean survival, months (95% CI) | Median survival, months (95% CI) | P value |
|---|---|---|---|---|---|---|
| Disease specific survival | ||||||
| Adenocarcinoma | Low (n=51) | 53.5 | 0.003 | 87.41 (68.91–195.90) | 80.00 (30.55–129.45) | 0.002 |
| High (n=51) | 26.7 | 49.24 (31.97–66.51) | 21.00 (15.52–26.48) | |||
| Squamous cell carcinoma | Low (n=54) | 46.6 | 0.156 | 80.53 (60.23–100.83) | 46.00 (1.13– 90.87) | 0.757 |
| High (n=57) | 59.2 | 73.89 (60.19–87.60) | 65.00 (44.30–85.70) | |||
| All | Low (n=105) | 50.1 | 0.461 | 81.71 (67.59–95.83) | 70.00 (40.69–99.31) | 0.348 |
| High (n=108) | 43.8 | 71.90 (57.59–86.22) | 41.00 (20.99–61.01) | |||
| Overall survival | ||||||
| Adenocarcinoma | Low (n=51) | 49.0 | <0.001 | 74.68 (58.27–91.09) | 59.00 (24.83–93.17) | 0.001 |
| High (n=51) | 21.6 | 39.31 (25.94–52.67) | 18.00 (14.12–21.88) | |||
| Squamous cell carcinoma | Low (n=54) | 36.9 | 0.355 | 65.27 (47.93–82.61) | 35.00 (21.80–48.20) | 0.590 |
| High (n=57) | 43.9 | 56.52 (45.55–67.48) | 55.00 (35.98–74.02) | |||
| All | Low (n=105) | 43.7 | 0.127 | 66.71 (55.15–78.28) | 47.00 (27.97–66.03 | 0.070 |
| High (n=108) | 32.4 | 53.36 (42.93–63.79) | 31.00 (21.60–40.40) |
CI, confidence interval; n, number; NHLRC2, NHL repeat (named after NCL-1, HT2A and LIN-41)-containing protein 2.
Semi-quantitative immunohistochemical NHLRC2 expression in cancer cells
To further study the NHLRC2 expression especially in cancer cells, the percentage of positive cancer cells was evaluated semi-quantitatively. There were more NHLRC2 positive cells in ADCs than in SCCs (P<0.001) since 50 (49%) out of 102 ADCs had over 75% positive cells while eighteen (16.2%) out of 111 SCCs had over 75% of positive cells (Table 3).
Table 3
| Cancer type | Negative, n (%) | <25% positive, n (%) | 25–49% positive, n (%) | 50–75% positive, n (%) | >75% positive, n (%) | Total, n (%) | P value |
|---|---|---|---|---|---|---|---|
| Adenocarcinoma | 2 (2.0) | 6 (5.9) | 18 (17.6) | 26 (25.5) | 50 (49.0) | 102 (100.0) | <0.001 |
| Squamous cell carcinoma | 1 (0.9) | 42 (37.8) | 28 (25.2) | 22 (19.8) | 18 (16.2) | 111 (100.0) | |
| Total | 3 (1.4) | 48 (22.5) | 46 (21.6) | 48 (22.5) | 68 (31.9) | 213 (100.0) |
NHLRC2, NHL repeat (named after NCL-1, HT2A and LIN-41)-containing protein 2.
NHLRC2 protein levels in tissue samples
To confirm the image analysis results of NHLRC2 expression, frozen ADC (n=3) and SCC (n=2) tissue samples were subjected to immunoblotting. NHLRC2 expression levels were variable in different tumor samples (Figure 4E, Figure S4), which is in line with the results of digital image analysis and semiquantitative analysis. NHLRC2 expression level was slightly higher in most of the tumor tissues compared to that in corresponding tumor-free lung but due to limited number of samples the result was not statistically significant.
NHLRC2 protein levels in cultured cells
NHLRC2 protein levels were measured in different types of cell lines by Western blot analysis to study whether there are differences in NHLRC2 expression between stromal cells cultured from lung cancer and areas outside tumor, normal epithelial cells of airways, and lung cancer cells in vitro. NHLRC2 protein levels were equal in stromal cells, normal epithelial cell lines (SAEC and PBTE) and epithelial cancer cell lines (H1650, SK-LU-1 and SK-MES-1) in the cell culture conditions (Figure 4F, Figure S5).
Discussion
As far as we are aware, this is the first study describing the immunohistochemical expression of NHLRC2 in lung tissue samples from patients with ADC and SCC. We observed that NHLRC2 protein and mRNA were mainly expressed in cancer cells and inflammatory cells within tumor stroma. We showed that immunohistochemical NHLRC2 expression in tumor tissue including all cell types assessed by image analysis method was higher in ADCs than in SCCs and it correlated with the short survival in ADC patients. Additionally, the proportion of NHLRC2-positive tumor cells was higher in ADCs than in SCCs.
The expression pattern of NHLRC2 in lung ADCs and SCCs in our study was in line to the pattern shown in the Human Protein Atlas with the same antibody (21,22). Human Protein Atlas presents negative (n=2) or weak (n=2) expression in SCC cancer cells (21). In ADCs, NHLRC2 expression was negative (n=1), weak or moderate in under 25% of the cells (n=2), or moderate in over 75% of the cells (n=3) (21). In normal lung NHLRC2 was reported to be expressed in epithelial cells of bronchi (23,24) as well as in type II pneumocytes and macrophages (24,25), which is also in line with our observations of the present study and the results of our previous study on IPF and normal lung (9).
Despite containing a Trx-like domain, NHLRC2 has not been shown to have Trx activity so far (5,26). The expression pattern of NHLRC2 in lung cancer tissues observed in the current study resembles that of Trx, since Trx has been detected in the cancer cells with varying extent (27-29). Additionally, also Trx expression has been reported in bronchial and alveolar epithelial cells and macrophages in normal lung outside tumor (27,30). Similar to NHLRC2 expression, Trx has been shown to be higher in tumor than in normal lung tissue by immunohistochemistry and Western blot analysis (29,31-33). Trx-like domain of NHLRC2 has been shown to be cleaved by caspase-8 in reactive oxygen species -induced apoptosis in human colon cancer cell line 8 (HCT116) (34). Different expression patterns of caspase-8 have been observed in lung ADC and SCC since ADC showed mainly diffuse cytoplasmic expression while strong cytoplasmic expression in single cells was dominant in SCC (35). Additionally, the single-cell staining pattern detected mainly in SCC was associated with high apoptotic activity (35).
In the current study, we showed that high NHLRC2 protein expression was associated with poor prognosis of ADC patients. According to the gene expression data presented in the Human Protein Atlas (21,22), NHLRC2 mRNA expression was not associated with survival in ADC or SCC although the grouping of patients was performed differently than we did. In contrast, a low gene expression of NHLRC2 combined with the expression of two other protein coding genes and one long non-coding RNA in tumor tissues has been reported to predict poor prognosis of lung ADC patients in a study using three gene expression datasets from Gene expression omnibus database (11). In that study, by combining protein-coding genes with long non-coding RNA, Ye and coauthors created a multidimensional transcriptome prognostic signature model that can predict survival probabilities in ADC patients (11). The apparent reasons for discrepant results of the study of Ye and ours is the fact that the methods used in Ye’s study (11) were very different than those of our study. Furthermore, it has been shown that mRNA and protein levels do not always correlate even in the same samples as seen in studies comparing data of proteomic and microarray analysis (36) and whole-genome sequencing, RNA sequencing and proteomics (37) performed on lung ADC tissues. That phenomenon may at least partly explain the different results between our study and those reported in Human Protein Atlas and by Ye and co-authors (11,21,22).
Positive immunohistochemical Trx expression has been associated with poor survival of lung cancer patients in some studies (30), but not all (27,28), when examinations were performed by using different semiquantitative scoring methods, and the histologic types were not taken into account (27,28,30). A higher proportion of Trx-positive samples was observed among SCCs than among ADCs in the study of Azuma (28), while Trx expression did not show association with histology in several other studies (27,30,33). The result was opposite to our finding regarding the different NHLRC2 expression in histologic lung cancer subtypes.
In the current study, large amount of clinical and histological data was collected and correlated with NHLRC2 expression. Of all clinical data and histopathological features analyzed only mitotic activity and survival were associated with the level of immunohistochemical NHLRC2 expression in ADC while no correlations were found between NHLRC2 expression and age, sex, smoking status, pulmonary function test results, COPD, growth pattern, nuclear atypia, tumor necrosis, desmoplasia, or lymphovascular invasion in ADC or SCC. Previously, micropapillary and solid predominant ADCs were shown to have worse 5-year DSS than other subtypes and high mitotic activity was associated with reduced DSS and OS in the same ADC cases used in the current study (13,15). Additionally, mucin-1 (MUC1) expression pattern has been evaluated in the same ADC cases revealing correlations with growth pattern, lymphovascular invasion, necrosis, and nuclear atypia (17). Mucin-1 has been shown to correlate also with stage and OS in ADC (17) while in the current study, NHLRC2 was associated with DSS and OS, but not with stage in the same patients. We observed that the NHLRC2 protein levels in different cell lines (including cancer cells from ADC and SCC) were nearly equal. Additionally, high immunohistochemical NHLRC2 expression was associated with high mitotic activity in ADC lung tissue samples, and thus it can be speculated whether NHLRC2 expression reflected the number of proliferating cells. When interpreting the results of cell line experiments, however, it is notable that each cell line was isolated and cultured from a tissue sample of one patient, and moreover, the gene expression may be altered during culture. Furthermore, the immunohistochemical NHLRC2 expression was determined in all cell types within tumors, not only in individual cell type as it has been done in cell lines.
This study was a retrospective analysis with a limited number of cases especially when the patients were divided into groups based on clinical and histological information. Due to the retrospective nature, some information was incomplete, which can be considered the weakness of the study. The collection of the patient’s clinical information and the histological re-analyses of the tumors have been, however, conducted in very detailed manner. Image analysis has several differences compared to the traditional semiquantitative analysis of immunohistochemical staining in different cell types. In digital image analysis software, total staining in all cell types in the sample is counted while in semiquantitative analyses different cell types can be scored individually. Based on our own experience, necrosis, anthracosis, variable sample quality, and background staining may be difficult to deal by the digital image analysis. On the other hand, the software can distinguish more reliably different intensities than human eye (38). Altogether, these two analysis methods have their own strengths and limitations, and thereby it may be advantageous to use them both.
Conclusions
NHLRC2 expression was higher in lung ADC than in SCC and its high expression was associated with mitotic activity and short survival in ADC patients. Further studies are required to clarify the pathogenetic role of NHLRC2 in lung cancer.
Acknowledgments
The authors thank Riitta Vuento from Center of Internal Medicine and Respiratory Medicine, Medical Research Center Oulu, Oulu University Hospital, and Research Unit of Biomedicine and Internal Medicine, University of Oulu for technical assistance with immunohistochemistry; Anne Heikkinen from ECM-Hypoxia Research Unit, University of Oulu; and Salla Kangas from Medical Research Center Oulu and Research Unit of Clinical Medicine, Oulu University Hospital and University of Oulu for designing the Visiopharm image analysis protocol used in this study. We also thank the Biocenter Oulu Transgenic and Tissue Phenotyping Core Facility, University of Oulu, Finland, a member of Biocenter Finland and Infrafrontier-EMMA.
Funding: This work was supported by the Foundation of the Finnish Anti-Tuberculosis Association (to MAK, and to RLK), the Tampere Tuberculosis foundation (to MAK), Väinö and Laina Kivi Foundation (to MAK), the Research Foundation of the Pulmonary Diseases (to MAK, and to RLK), Orion research foundation (to MAK), Medical Research Center Oulu (to STL), Academy of Finland profiling program (Decision No. 311934, to RMLH), University of Oulu (to RMLH), the Foundation for Pediatric Research, Finland (to RMLH), the Jalmari and Rauha Ahokas Foundation (to RLK), the Research Foundation of the North Finland (to RLK), and the state subsidy of Oulu University Hospital (to RLK). Funding sources were not involved in study design, collection, analysis and interpretation of data, writing of the report, or decision to submit the article for publication.
Footnote
Reporting Checklist: The authors have completed the RECORD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-815/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-815/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-815/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-815/coif). MAK reports personal grants for scientific work from the Finnish Anti-Tuberculosis Association, the Tampere Tuberculosis foundation, Väinö and Laina Kivi Foundation, the Research Foundation of the Pulmonary Diseases, and Orion research foundation. STL reports the research grant received from MRC Oulu (Medical Research Center Oulu). HELB reports speaker fees from Roche, Amgen Aktiebolag and Boehringer Ingelheim; support for attending training course from MSC Finland; stocks from Orion Oyj. RMLH reports the research grant received from the Foundation for Pediatric Research and support from Academy of Finland and the University of Oulu. RLK reports grants for the study group from the state subsidy of Oulu University Hospital, the Tampere Tuberculosis Foundation, the Jalmari and Rauha Ahokas Foundation, the Research Foundation of the North Finland, The Finnish-Anti-Tuberculosis Association Foundation, and the Research Foundation of the Pulmonary Diseases; consulting fees from Boehringer Ingelheim and MSD; lecture fees from Boehringer Ingelheim and Roche; virtual congress cost from Roche and Novartis; being the President of the Finnish Respiratory Society between March 2017 and March 2020; being member of the board of the Finnish Medical Foundation between January 2020 and December 2022 and member of the board of Finnish Lung Health association from January 2021 onwards. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). A favorable statement of the study protocol was given by the Ethical Committee of Northern Ostrobothnia Hospital District in Oulu (2/2008, amendments 12/2014, 2/2015, 2/2018 and 6/2022). National Supervisory Authority for Welfare and Health has approved the research use of paraffin-embedded tissue samples (Dnro: V/25090/2019) and individual consent for this retrospective analysis was waived. For collection of cell culture materials, all subjects gave their written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- WHO Classification of Tumours Editorial Board. Thoracic Tumours, In: WHO Classification of Tumours series. 5th ed. Lyon, France: International Agency for Research on Cancer; 2021.
- Yang Y, Wang M, Liu B. Exploring and comparing of the gene expression and methylation differences between lung adenocarcinoma and squamous cell carcinoma. J Cell Physiol 2019;234:4454-9. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Uusimaa J, Kaarteenaho R, Paakkola T, et al. NHLRC2 variants identified in patients with fibrosis, neurodegeneration, and cerebral angiomatosis (FINCA): characterisation of a novel cerebropulmonary disease. Acta Neuropathol 2018;135:727-42. [Crossref] [PubMed]
- Brodsky NN, Boyarchuk O, Kovalchuk T, et al. Novel compound heterozygous variants in NHLRC2 in a patient with FINCA syndrome. J Hum Genet 2020;65:911-5. [Crossref] [PubMed]
- Rapp CK, Van Dijck I, Laugwitz L, et al. Expanding the phenotypic spectrum of FINCA (fibrosis, neurodegeneration, and cerebral angiomatosis) syndrome beyond infancy. Clin Genet 2021;100:453-61. [Crossref] [PubMed]
- Hiltunen AE, Vuolteenaho R, Ronkainen VP, et al. Nhlrc2 is crucial during mouse gastrulation. Genesis 2022;60:e23470. [Crossref] [PubMed]
- Kreus M, Lehtonen S, Hinttala R, et al. NHLRC2 expression is increased in idiopathic pulmonary fibrosis. Respir Res 2022;23:206. [Crossref] [PubMed]
- Boon K, Bailey NW, Yang J, et al. Molecular phenotypes distinguish patients with relatively stable from progressive idiopathic pulmonary fibrosis (IPF). PLoS One 2009;4:e5134. [Crossref] [PubMed]
- Ye J, Liu H, Xu ZL, et al. Identification of a multidimensional transcriptome prognostic signature for lung adenocarcinoma. J Clin Lab Anal 2019;33:e22990. [Crossref] [PubMed]
- Kreus M, Lehtonen S, Skarp S, et al. Extracellular matrix proteins produced by stromal cells in idiopathic pulmonary fibrosis and lung adenocarcinoma. PLoS One 2021;16:e0250109. [Crossref] [PubMed]
- Mäkinen JM, Laitakari K, Johnson S, et al. Nonpredominant lepidic pattern correlates with better outcome in invasive lung adenocarcinoma. Lung Cancer 2015;90:568-74. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke A, et al. WHO classification of tumours of the lung, pleura, thymus and heart. 4th Edition. Lyon, France: International Agency for Research on Cancer; 2015.
- Mäkinen JM, Laitakari K, Johnson S, et al. Histological features of malignancy correlate with growth patterns and patient outcome in lung adenocarcinoma. Histopathology 2017;71:425-36. [Crossref] [PubMed]
- Rusch VW, Appelman HD, Blackstone E, et al. AJCC Cancer Staging Handbook from the AJCC Cancer Staging Manual. In: Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. Springer, 2010. pp. 299–323.
- Lappi-Blanco E, Mäkinen JM, Lehtonen S, et al. Mucin-1 correlates with survival, smoking status, and growth patterns in lung adenocarcinoma. Tumour Biol 2016;37:13811-20. [Crossref] [PubMed]
- Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22-9. [Crossref] [PubMed]
- Karvonen HM, Lehtonen ST, Sormunen RT, et al. Myofibroblasts in interstitial lung diseases show diverse electron microscopic and invasive features. Lab Invest 2012;92:1270-84. [Crossref] [PubMed]
- Karvonen HM, Lehtonen ST, Sormunen RT, et al. Lung cancer-associated myofibroblasts reveal distinctive ultrastructure and function. J Thorac Oncol 2014;9:664-74. [Crossref] [PubMed]
- Human Protein Atlas. Expression of NHLRC2 in lung cancer - The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000196865-NHLRC2/pathology/lung+cancer (accessed 15 September 2022).
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [Crossref] [PubMed]
- Tissue expression of NHLRC2 - Staining in bronchus - The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000196865-NHLRC2/tissue/bronchus (accessed 15 September 2022).
- Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Tissue expression of NHLRC2 - Staining in lung - The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000196865-NHLRC2/tissue/lung (accessed 15 September 2022).
- Yeung ATY, Choi YH, Lee AHY, et al. A Genome-Wide Knockout Screen in Human Macrophages Identified Host Factors Modulating Salmonella Infection. mBio 2019;10:e02169-19. [Crossref] [PubMed]
- Soini Y, Kahlos K, Näpänkangas U, et al. Widespread expression of thioredoxin and thioredoxin reductase in non-small cell lung carcinoma. Clin Cancer Res 2001;7:1750-7. [PubMed]
- Azuma K, Komohara Y, Sasada T, et al. Excision repair cross-complementation group 1 predicts progression-free and overall survival in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Sci 2007;98:1336-43. [Crossref] [PubMed]
- Fernandes AP, Capitanio A, Selenius M, et al. Expression profiles of thioredoxin family proteins in human lung cancer tissue: correlation with proliferation and differentiation. Histopathology 2009;55:313-20. [Crossref] [PubMed]
- Kakolyris S, Giatromanolaki A, Koukourakis M, et al. Thioredoxin expression is associated with lymph node status and prognosis in early operable non-small cell lung cancer. Clin Cancer Res 2001;7:3087-91. [PubMed]
- Kim HJ, Chae HZ, Kim YJ, et al. Preferential elevation of Prx I and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell Biol Toxicol 2003;19:285-98. [Crossref] [PubMed]
- Park JH, Kim YS, Lee HL, et al. Expression of peroxiredoxin and thioredoxin in human lung cancer and paired normal lung. Respirology 2006;11:269-75. [Crossref] [PubMed]
- Deng ZH, Cao HQ, Hu YB, et al. TRX is up-regulated by fibroblast growth factor-2 in lung carcinoma. APMIS 2011;119:57-65. [Crossref] [PubMed]
- Nishi K, Iwaihara Y, Tsunoda T, et al. ROS-induced cleavage of NHLRC2 by caspase-8 leads to apoptotic cell death in the HCT116 human colon cancer cell line. Cell Death Dis 2017;8:3218. [Crossref] [PubMed]
- Törmänen-Näpänkangas U, Soini Y, Kahlos K, et al. Expression of caspases-3, -6 and -8 and their relation to apoptosis in non-small cell lung carcinoma. Int J Cancer 2001;93:192-8. [Crossref] [PubMed]
- Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 2002;1:304-13. [Crossref] [PubMed]
- Soltis AR, Bateman NW, Liu J, et al. Proteogenomic analysis of lung adenocarcinoma reveals tumor heterogeneity, survival determinants, and therapeutically relevant pathways. Cell Rep Med 2022;3:100819. [Crossref] [PubMed]
- Lykkegaard Andersen N, Brügmann A, Lelkaitis G, et al. Virtual Double Staining: A Digital Approach to Immunohistochemical Quantification of Estrogen Receptor Protein in Breast Carcinoma Specimens. Appl Immunohistochem Mol Morphol 2018;26:620-6. [Crossref] [PubMed]


