
Biomarker analysis of the phase II JO25567 study comparing erlotinib with or without bevacizumab in first-line advanced EGFR+non-small-cell lung cancer
Highlight box
Key findings
• In this exploratory biomarker analyses of the phase II JO25567 study, EB showed favorable treatment outcomes in patients with low pVEGFA and serum leptin, and limited efficacy in patients with high serum follistatin.
What is known and what is new?
• The JO25567 study found that EB significantly prolonged median PFS compared with E, without unexpected adverse effects; HR 0.54 (95% CI: 0.36–0.79).
• A consistent predictive biomarker for bevacizumab treatment has not yet been validated across any cancer type. This study is thought to be advantageous for biomarker analyses for bevacizumab due to the relatively homogeneous patient population selected.
What is the implication, and what should change now?
• We expect our results will provide a basis for validation studies and contribute toward understanding the therapeutic positioning of the combination therapy among various solutions to improve EGFR-TKI outcomes for patients with EGFR-positive NSCLC.
Introduction
Lung cancer is a leading cause of cancer-related death worldwide, and a significant proportion of patients with non-small-cell lung cancer (NSCLC) have activating mutations in the epidermal growth factor receptor (EGFR) gene (1,2). First-generation EGFR-tyrosine kinase inhibitors (TKIs), such as erlotinib, were the standard of care for patients with activating EGFR mutation-positive NSCLC (3-5). The third-generation EGFR-TKI, osimertinib has demonstrated improved progression-free survival (PFS) and overall survival (OS) compared with the first-generation EGFR-TKIs, erlotinib and gefitinib (6,7). However, most patients experience disease progression during their treatment course with EGFR-TKIs. New strategies may overcome development of resistance.
Bevacizumab is a humanized monoclonal antibody against vascular endothelial growth factor-A (VEGF-A), and has been shown to provide additional efficacy when used in combination with chemotherapy in multiple tumor types (8,9). Bevacizumab inhibits VEGF-induced neovascularization needed for tumor growth, and partially normalizes tumor vessels with abnormal, hyperpermeable properties caused by VEGF-A, thereby improving the interstitial fluid pressure of the tumor (10).
EGFR-TKIs are thought to inhibit VEGF-A activity through cross-talk between their respective signaling pathways (11). In preclinical studies, combined VEGF and EGFR inhibiton was found to postpone the development of EGFR-TKI resistance, and was still effective in EGFR-TKI-resistant tumors (12,13). Moreover, in the phase II JO25567 study, erlotinib plus bevacizumab (EB) provided a 6.3-month extension in median PFS compared with erlotinib alone (E) in patients with advanced EGFR mutation-positive non-squamous NSCLC (14). EB significantly prolonged median PFS {16.0 [95% confidence interval (CI): 13.9–18.1] months} compared with E [9.7 (95% CI: 5.7–11.1) months], without unexpected adverse effects; hazard ratio (HR) 0.54 (95% CI: 0.36–0.79; P=0.0015). To understand the additional effect of EB, we analyzed biomarkers that are thought to correlate with EB efficacy.
Plasma VEGF-A (pVEGFA) is one of the most frequently investigated biomarkers for bevacizumab. The phase III studies, AVAGAST, AVADO, and AViTA, demonstrated that high pVEGFA levels correlate with longer PFS and/or OS in patients with advanced gastric cancer (GC), metastatic breast cancer (mBC), and pancreatic cancer (PAC) treated with bevacizumab (15-17). However, this correlation has not been consistently replicated across different studies and cancer types (18). Various circulating angiogenic factors (CAFs) other than VEGF-A have also been analyzed for their association with bevacizumab treatment outcomes; soluble VEGF receptor 1 (sVEGFR1), interleukin-8 (IL-8), and angiopoietin 2 were reported to have some predictive potential, but not for patients with NSCLC (19).
Tumor tissue expression levels of VEGF pathway genes and proteins have been studied to find biomarkers for bevacizumab, but few correlations have been demonstrated. VEGF-A gene expression has been suggested as a surrogate marker for angiogenesis (20). Recent studies suggest the importance of separate evaluation of the VEGF-A splice isoforms, VEGF-Axxxa and xxxb, to predict bevacizumab treatment outcomes in patients with BC and colorectal cancer (CRC). Although bevacizumab binds both isoforms, VEGF-Axxxb has an anti-angiogenic profile that differs from pro-angiogenic VEGF-Axxxa (21,22). Low tumor tissue expression levels of baseline VEGFR1 and co-receptor neuropilin 1 have been reported to correlate with better bevacizumab treatment outcomes in several phase III studies in patients with GC, CRC, and mBC (15,19,23).
Tumor endothelial cells (ECs) are the main target of VEGF-A; therefore, the tumor angiogenic response to bevacizumab treatment is thought to be influenced by polymorphisms in angiogenesis-related genes. Several studies have reported that polymorphisms in angiogenesis-related genes affect PFS, OS, and/or severe hypertension in bevacizumab-treated patients with various cancer types, including NSCLC, renal-cell carcinoma, mBC, and PAC (19,24).
Although several biomarkers for bevacizumab treatment efficacy look promising, a consistent predictive biomarker has not been validated across any cancer type. Since NSCLC is categorized into various subtypes based on genotype and histology, biomarker analyses may also be needed within NSCLC subtypes. Therefore, the JO25567 study is thought to be advantageous for biomarker analyses for bevacizumab due to the relatively homogeneous patient population selected (Japanese patients with previously untreated EGFR-positive non-squamous NSCLC). In the present study, we performed biomarker analyses for EB treatment using data from the JO25667 study, aiming to: (I) confirm the association between bevacizumab treatment outcomes and pVEGFA level; (II) screen possible predictive biomarker candidates for bevacizumab, including CAFs, messenger RNAs (mRNAs) and proteins in tumor tissues, and polymorphisms in angiogenesis-related genes; (III) understand the underlying mechanisms of bevacizumab efficacy when given as EB. We present this article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-632/rc).
Methods
Study design and patients
The study design and patient population for the JO25567 study have been reported previously (14). Briefly, JO25567 was a multicenter, randomized, open-label, phase II study examining the addition of EB as a first-line therapy in patients with NSCLC harboring activating EGFR mutations. Between February 21, 2011 and March 5, 2012, 154 patients were enrolled from 30 centers in Japan and randomly assigned 1:1 to receive either erlotinib 150 mg once daily plus bevacizumab 15 mg/kg every 3 weeks or erlotinib 150 mg once daily. The primary endpoint was PFS determined by an independent review committee. Secondary endpoints were OS, tumor response, quality of life, and safety. An exploratory objective was to seek predictive biomarkers for EB treatment outcomes.
Eligible patients had histologically and/or cytologically confirmed stage IIIB/IV or postoperative recurrent non-squamous NSCLC with an activating EGFR mutation (either exon 19 deletion or L858R mutation). Key exclusion criteria included confirmation of T790M mutation and the presence of brain metastases. Detailed inclusion and exclusion criteria have been published (14).
The study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and Good Clinical Practice guidelines. The study protocol was reviewed and approved by the institutional review boards of the participating institutions, and written informed consent was obtained from all patients. The study was registered with the Japan Pharmaceutical Information Center (No. JapicCTI-111390).
Procedures for biomarker analyses
Informed consent was obtained from patients before sample collection for each type of biomarker analysis. Five types of biomarker were analyzed (Figure 1): pVEGFA, CAFs in serum, polymorphisms of angiogenesis-related genes, and mRNAs and proteins in tumor tissue specimens.
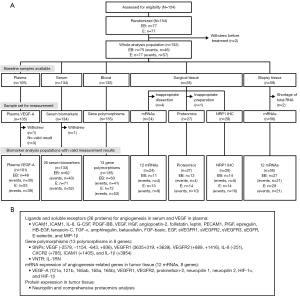
Tumor tissue specimens were removed at diagnosis or during surgery and the formalin-fixed and paraffin-embedded (FFPE) samples were stored at room temperature. Blood samples for the analysis of CAFs were taken before the initiation of the first treatment administration; ethylenediamine tetraacetic acid (EDTA) plasma and serum were prepared and stored frozen at −70 ℃ before analysis. For genotyping, a 5 mL blood sample was taken from each patient between day 1 of treatment cycle 1 and the final visit, and stored below −70 ℃.
pVEGFA
pVEGFA was analyzed in duplicate by multiplex enzyme-linked immunosorbent assay (ELISA) methods using Immunological Multiparametric Chip Technique (IMPACT) technology (Roche Diagnostics GmbH, Mannheim, Germany) (18,25). The analysis was performed according to User Guidelines ONC 2.03 version 02 for IMPACT by Roche Diagnostics GmbH.
Quantification of ligands and soluble receptors in serum
The concentrations of secreted proteins in serum samples were determined using commercially available bead-based sandwich immunoassay kits. Vascular cell adhesion molecule 1, intercellular adhesion molecule 1 (ICAM1), macrophage inflammatory protein-1β (MIP-1β), and E-selectin were assayed using the Procarta Cytokine Assay Kit (PN-PC1002) (Panomics, Inc., Fremont, CA, USA). IL-8, granulocyte colony-stimulating factor (G-CSF), VEGF, platelet-derived growth factor BB (PDGF-BB), hepatocyte growth factor (HGF), angiopoietin-2, follistatin, leptin, and platelet/endothelial cell adhesion molecule 1 (PECAM-1) were determined using the Bio-Plex Pro Human Angiogenesis Assay Panel (Bio-Rad Laboratories, 171-A4011M, CA, USA). Placental growth factor (PlGF), epiregulin, heparin-binding epidermal growth factor-like growth factor (HB-EGF), tenascin-C, transforming growth factor (TGF)-α, amphiregulin, betacellulin, basic fibroblast growth factor (bFGF), and EGF were measured using the WideScreen Human Cancer Panel 2 (BPHCPP002-BASE) (Merck, Kenilworth, NJ, USA). Levels of soluble receptors sVEGFR-1, sVEGFR-2, sVEGFR-3, and sEGFR were determined using the Human Soluble Cytokine Receptor Panel 4-plex (HSCR-32K 4-plex) (Millipore, St. Charles, MO, USA). All assays were performed according to each manufacturer’s instructions. Measurements were run using a Bio-Plex Suspension Array Full System (Bio-Rad, Hercules, CA, USA).
Genotyping
Key polymorphisms in angiogenesis-related genes were analyzed by LSI Medience Co. (Tokyo, Japan). Genes were extracted from blood samples and amplified by polymerase chain reaction (PCR) using the GeneAmp® PCR System 9700 Dual 96 Well system (Applied Biosystems®, Foster City, CA, USA). A total of 12 single nucleotide polymorphisms (SNPs) were detected by standard PCR reaction and DNA direct sequencing methods in seven genes [VEGF, VEGFR1, VEGFR2, IL-8, CXC chemokine receptor 2 (CXCR2), ICAM1, and IL-1β], using a 3130xl Genetic Analyzer (Applied Biosystems®, Foster City, CA, USA). A variable number tandem repeat (VNTR) was analyzed by electrophoresis in the IL-1 receptor antagonist protein (IL-1RN) using the Microchip Electrophoresis System for DNA/RNA Analysis MCE®-202 MultiNA (Shimadzu, Kyoto, Japan).
Gene expression analysis by MassARRAY
Total RNA was isolated from FFPE slides of tumor tissue and analyzed using the multiplex analysis of gene expression system, MassARRAY (LungCarta Panel; Sequenom, San Diego, CA, USA). Specifically, the FFPE specimens were divided into sections (4 µm thick), which were placed on slides and deparaffinized. For surgical specimens, one of the serial sections was stained with hematoxylin and eosin as a reference slide. The tumor-rich area of the reference slide was marked by a pathologist. Guided by the slide, the corresponding tumor-rich area was manually dissected in the remaining serial sections to avoid contamination of non-tumor tissue (macrodissection). The macrodissection procedure was not applied for biopsy specimens. Total RNA was extracted, purified, and measured quantitatively using the Quanti-iTTM RiboGreen® RNA Assay Kit (Life Technologies Japan Ltd., Tokyo, Japan) according to the manufacturer’s instructions. mRNAs of VEGF-A, VEGFR1, VEGFR2, prokineticin-2, neuropilin 1, neuropilin 2, hypoxia-inducible factor-1α (HIF-1α), and HIF-1β were analyzed via MassARRAY in reference to beta-actin (ACTB), hypoxanthine phosphoribosyltransferase-1 (HPRT1), ribosomal protein L13A (RPL13A), and 14-3-3 protein zeta/delta (YWHAZ) housekeeping genes, which were used as internal standards.
Immunohistochemistry (IHC) for neuropilin-1
IHC was performed on FFPE slides of tumor tissue at LSI Medience Co. (Tokyo, Japan). The human neuropilin-1 antibody [monoclonal mouse immunoglobulin G1 clone #446915, Code No. MAB38701] was purchased from R&D Systems, Inc. Reference tumor tissue slides of human colon cancer, adenocarcinoma, grade I (Code No. HuCAT116), and human colon cancer, adenocarcinoma, grade II (Code No. HuCAT121) were purchased from US BioMax, Inc. and stored at 4 ℃.
Serial sections were prepared; one section was stained with hematoxylin and eosin and one section was used for IHC with the human neuropilin-1 antibody. The intensity of staining in each tumor cell was assessed among a total of 500 cells. An H-score was calculated as an aggregate of the percentage of stained tumor cells at each intensity for a score between 0 and 3 as follows: H score = (0x%) + (1x%) + (2x%) + (3x%), where 0, 1, 2, and 3 were negative, weak, intermediate, and strong staining, respectively.
Statistical analysis
The modified intent-to-treat population for the efficacy analysis included all patients who had received at least one dose of study treatment and had a tumor assessment at least once after randomization. There were no preplanned sample size or power calculations for the biomarker analyses. Median PFS in each subgroup was estimated using the Kaplan-Meier method; Greenwood’s formula was used to calculate 95% CIs.
pVEGFA levels were categorized into high and low using the median as a cutpoint. To assess the predictive value of each biomarker, Cox proportional hazards models were used to determine HRs and 95% CIs with the following terms: EB treatment, biomarker status, and interaction between EB treatment and biomarker status, with or without adjustment by the stratification factors. Patients were stratified according to sex (men vs. women), disease stage (stage IIIB vs. stage IV vs. postoperative relapse), smoking history (never smokers or former light smokers vs. others), and type of EGFR mutation (exon 19 deletion vs. L858R mutation). Multiplicities were not considered in the analyses. Interaction of continuous variable predictors (categorized using median or quartile cutpoints) with treatment was analyzed. Secondarily, the multivariable fractional polynomial interaction (MFPI) procedure and tail-oriented subpopulation treatment effect pattern plot (TO-STEPP) were applied following Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines (26-28).
The statistical analysis methodology for gene polymorphisms is provided in the Appendix 1. All analyses were conducted using Statistical Analysis Systems, version 9.2 (SAS Institute, Cary, NC, USA) and R version 3.0.3.
Results
Patients and samples
Of the 154 enrolled patients, the data from 152 patients (EB, n=75; E, n=77) were included in the analysis population (Figure 1). At the data cutoff date (30 June 2013), 103 progression events had occurred; median follow-up was 20.4 months (14).
The baseline characteristics of patients in each biomarker-analysis population were generally similar to the whole analysis population with respect to age, gender, disease stage, smoking history, and type of EGFR mutation (except for those with surgical tissue specimens), and well balanced between treatment groups in each analysis population (Table S1). However, the proportion of patients with recurrent disease was higher in the surgical tissue sample population than the whole analysis population (Table S1).
pVEGFA
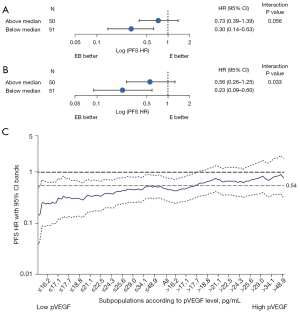
We evaluated the predictive value of pVEGFA for EB treatment efficacy in 105 EDTA plasma samples. Median pVEGFA levels were 18.0 pg/mL [minimum (min), 7.7 pg/mL; maximum (max), 95.8 pg/mL] in the EB treatment group and 18.8 pg/mL (min, 9.1 pg/mL; max, 59.2 pg/mL) in the E group.
On Cox analysis, pVEGFA levels were associated with EB treatment outcomes, suggesting that pVEGFA levels may have predictive value (Figure 2A,2B). EB treatment improved PFS vs. E alone for patients with pVEGFA levels below the median in patients with (HR, 0.23; 95% CI: 0.09–0.60) or without (HR, 0.30; 95% CI: 0.14–0.63) adjustment for stratification factors. The risk was reduced by 44% (HR, 0.56; 95% CI: 0.26–1.25) or 27% (HR, 0.73; 95% CI: 0.39–1.39) in patients with pVEGFA levels above the median with or without adjustment for stratification factors, respectively. The interaction P values were 0.033 between EB treatment outcomes and pVEGFA levels with adjustment for stratification factors. The interaction was visualized using TO-STEPP methodology, which demonstrated that the HR was positively related to pVEGFA levels assessed as a continuous variable (Figure 2C) (26).
Serum ligands and soluble receptors
We analyzed 26 ligands and soluble receptors in 134 serum samples available among the 152 patients included from JO25567. Cox regression analysis was performed for the interaction of EB treatment outcomes (PFS) with four ligand/receptor subgroups categorized by quartile cutpoints of the biomarker levels at primary screening. We screened one soluble receptor (sVEGFR3) and four ligands (follistatin, leptin, IL-8, and MIP-1β) as candidate biomarkers, with an interaction P value <0.2. HRs (EB vs. E treatment) for PFS in subgroups by biomarker quartiles and interaction P values are shown in Figure 3.
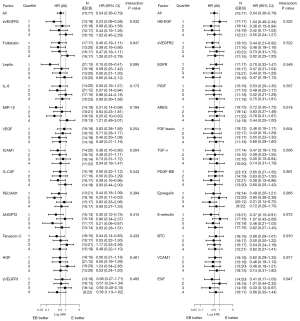
Secondly, we evaluated interactions between treatment outcomes and biomarkers using the MFPI approach (27). Table 1 shows the interaction between EB treatment outcomes and follistatin, leptin, and MIP-1β levels, with P<0.05 without adjustment for stratification factors. When four stratification factors were incorporated in the MFPI analysis as covariates (gender, disease stage, smoking history, and type of EGFR mutation), follistatin and leptin levels also had significant interactions (P<0.05) with EB treatment outcomes, but MIP-1β did not. Their relationships were visualized using TO-STEPP methodology (Figure 4A,4B) (26).
Table 1
| Biomarker | Model† | P value of interaction | |
|---|---|---|---|
| Without covariate | With covariate | ||
| Leptin | αX−2 | 0.0037 | 0.0049 |
| Follistatin | αX3 | 0.0148 | 0.0168 |
| MIP-1β | αX0.5 | 0.0459 | 0.0548 |
| sVEGFR3 | αlog(X) | 0.0586 | 0.0593 |
| IL-8 | αX−2 + βX−2 log(X) | 0.3142 | 0.1717 |
†, model chosen by fractional polynomial methodology. MIP-1β, macrophage inflammatory protein-1β; sVEGFR3, soluble vascular endothelial growth factor receptor 3; IL-8, interleukin 8.
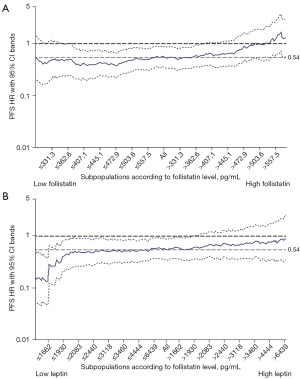
Cutpoints [by methods according to Jiang et al. (29)] were estimated at 490.5 (95% CI: 251.6–563.0) and 1,662.4 (95% CI: 1,245.1–5,966.8) pg/mL for follistatin and leptin, respectively, and were supported by STEPP (Figure 4A,4B). Among the 133 patients analyzed, 35 were above the cutpoint for follistatin and 40 were below the cutpoint for leptin. Even in patients with higher leptin levels (n=93), median PFS was still longer in the EB-treated group [16.0 (95% CI: 13.4–20.9) months] than the E-treated group [11.1 (95% CI: 8.0–18.0) months].
Univariate logistic regression analysis revealed a significant correlation between follistatin levels dichotomized at its cutpoint (490.5 pg/mL) and baseline serum concentrations of 12 out of 25 evaluated serum angiogenesis-related proteins. PECAM-1, G-CSF, IL-8, HGF, HB-EGF, amphiregulin, betacellulin, bFGF, tenascin-C, angiopoietin 2, epiregulin, and PlGF were significant under the threshold q value <0.05 [false discovery rate (FDR) by Benjamini and Hochberg (30)]. Figure S1 shows the distribution of serum concentrations of the 12 proteins divided into two subgroups of patients who had follistatin levels above and below the cutpoint.
Gene expression in tissue specimens
Tumor tissue specimens were collected from 82 patients (58 by biopsy; 24 surgically). MassARRAY analysis for mRNAs of angiogenesis-related genes was performed using the extracted RNA, excluding two biopsy samples due to a shortage of total RNA (<10 ng) extracted. For the statistical analysis, data were used from the 24 surgical samples only, since contamination of mRNA was suspected in the biopsy samples from peripheral normal tissues but not tumor tissues (data not shown). Further analysis for prokineticin-2 was not performed, as the mRNA level was only determined by MassARRAY in 2/24 samples.
Results of the interaction between EB treatment outcomes and mRNA expression levels of VEGF-A isoforms and other angiogenesis-related genes using Cox regression analysis are shown in Figure 5A. VEGF-A165a mRNA was suggested to interact with EB treatment outcomes (P=0.034), but no interactions were observed in the other mRNAs analyzed. Patients with VEGF-A165a mRNA levels below the median seemed sensitive to EB treatment efficacy.
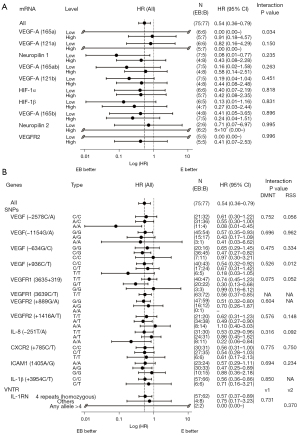
Gene polymorphisms
A total of 135 blood samples (from 152 patients) were available for genotype analysis of angiogenesis-related genes. We analyzed 12 SNPs and one VNTR of eight genes including VEGF-A, VEGFR1, VEGFR2, CXCR2, ICAM1, IL-1β, IL-8, and IL-1RN.
Figure 5B shows the HRs (EB vs. E) for PFS in the genotypes analyzed. Patients who were homozygous with the minor allele VEGF-A +936C/T (rs3025039) SNP T/T were suggested to have a favorable median PFS with EB treatment compared with those with C/T or C/C; the interaction of its recessive effect with EB treatment was P=0.012 (but only 11 patients had T/T). The minor alleles of VEGF-A −2578A/C (rs699947) (A/A, n=15) and VEGFR1 rs9582036 (G/G, n=6) showed a marginal recessive effect, with P=0.056 and 0.052, respectively (Figure 5B). The G allele of VEGFR1 3635+319G/T (rs9582036) also showed a marginal dominant effect with P=0.075, but the direction of shift in HR with T/G from major alleles T/T was different from those with G/G (Figure 5B). All 135 patients had a T/T genotype in VEGFR1 +3639C/T (rs7993418). No other significant interactions were observed between EB treatment outcomes and the gene polymorphisms analyzed.
Protein levels in tumor tissues
Neuropilin was evaluated in tumor tissue (n=28) by IHC but no interaction was detected; interaction P values were 0.997 or 0.858 in Cox regression models with or without adjustment for the four stratification factors, where neuropilin H-score levels were categorized as below (n=14) or above the median (n=14). Proteomic analysis was planned but could not be performed due to a shortage of tissue samples for analysis.
Discussion
JO25567 was the first prospective randomized study that demonstrated significant prolongation of PFS by combining bevacizumab with an EGFR-TKI in patients with EGFR-mutation positive non-squamous NSCLC. In the current biomarker analysis from JO25567, we observed that baseline levels of pVEGFA, serum follistatin, and leptin were associated with bevacizumab treatment efficacy, when given in combination with erlotinib.
In the current study, patients with pVEGFA levels below the median were considered sensitive to EB treatment; however, this is inconsistent with previous reports showing that higher pVEGFA levels led to better outcomes following bevacizumab treatment in patients with mBC, GC, and PAC (15-17). It is notable that median pVEGFA levels were 18.0 pg/mL in the EB-treated group and 18.8 pg/mL in the E-treated group in the JO25567 study; these are much lower than previous studies. Median pVEGFA was 125 pg/mL in patients with mBC in AVADO and 111 pg/mL in patients with GC in AVAGAST, even though pVEGFA was measured by the same IMPACT assay, which is sensitive to short isoforms (VEGF-A111 and 121) (15,16). Miles et al. reported that baseline pVEGFA levels were not predictive of a PFS benefit from bevacizumab in the MERiDiAN phase III trial, which prospectively investigated the correlation of pVEGFA and bevacizumab efficacy in patients with HER2-negative mBC (31). Additionally, a comprehensive reassessment of 12 clinical studies of eight tumors suggested that pVEGFA is not a robust predictive marker of bevacizumab treatment outcomes (18). On the other hand, Bai et al. reported a similar trend to the current findings in patients with mCRC, with patients with lower pVEGF-A121 levels gaining more benefit in terms of PFS and OS following bevacizumab treatment (32). They discussed that the roles of VEGF-A121 may be context- and cancer type-dependent. In the case of the current study, the analysis may be affected by the relatively restricted population of Japanese patients who had genetically and histologically homogeneous tumors [non-squamous EGFR active mutation-positive NSCLC (mainly adenocarcinoma)]. Further study will be needed in this population.
In the current study, gene expression analysis in tumor tissues showed that patients with lower VEGF-A165a mRNA levels seemed to be sensitive to EB treatment, though the sample size was small (n=24), further supporting pVEGFA as a potent predictive biomarker for bevacizumab treatment outcomes, at least in Japanese patients with NSCLC harboring an EGFR mutation. However, Pentheroudakis et al. reported that a higher level of VEGF-Axxxa mRNA in tumor tissue was associated with adverse prognosis in the absence of bevacizumab but with favorable outcomes when bevacizumab was administered in patients with advanced BC (22). This discrepancy may be derived from the genetically and histologically homogeneous tumor profile in the JO25567 study, as mentioned above.
EB treatment efficacy was reduced in patients with higher serum follistatin levels. The primary function of follistatin, a 31–39 kDa glycosylated cysteine-rich polypeptide, involves binding and neutralizing some members of the TGF-α superfamily, such as activin A (ActA) and myostatin. The interaction between follistatin and ActA results in an almost irreversible binding complex that prevents ActA from interacting with its receptors. Follistatin, widely expressed in various human tissues, is a product of a single gene on chromosome 5q11.2, which is translated into at least two molecular forms by alternative splicing and post-translational modifications i.e., follistatin 288 and 315. The latter is the predominant circulating form, existing mainly in a bound state (33).
Chen et al. reported that: (I) elevated follistatin in serum from patients with lung adenocarcinoma is directly produced by tumor cells; (II) serum follistatin is higher in patients with poorly-differentiated adenocarcinoma than in those with well-differentiated tumors; and (III) follistatin may promote the survival of lung adenocarcinoma cells by inhibiting apoptosis (34). Gökmen-Polar et al. reported a suggestive result in xenograft models, with the follistatin pathway being one of the top signaling pathways associated with resistance to bevacizumab in a human breast cancer cell line (MCF-7) over expressing the VEGF-A gene (35).
Many investigators have reported that the follistatin/ActA system plays an important role in neovascularization but their findings are controversial. Some have reported that constitutively expressed ActA in ECs inhibits EC proliferation in an autocrine or paracrine manner, to maintain a quiescent endothelium, and that follistatin stimulates EC proliferation and angiogenesis by neutralizing the growth inhibitory effect of ActA (36,37). Conversely, an anti-angiogenic effect of follistatin has also been reported: ActA has been shown to increase the expression of VEGF-A, VEGFR1, and VEGFR2 in an autocrine or paracrine manner, and follistatin reduced the expression of VEGFR1 and VEGFR2 by blocking ActA (38). In addition, ActA induces apoptosis in various tumor cells, and follistatin blocks its activity (38,39). Combining these findings, follistatin is speculated to inhibit VEGF-A-dependent angiogenesis, but stimulate neovascularization independently from VEGF-A/VEGFRs and promote the survival of tumor cells. This speculation may explain the limited efficacy of bevacizumab in patients with high serum follistatin in the present study.
The production of multiple growth factors, including angiogenic factors, has been reported in NSCLC cells (40,41). Frezzetti et al. reported that VEGF-A induced the secretion of a variety of angiogenic factors, including follistatin, HGF, angiopoietin-2, G-CSF, IL-8, leptin, PECAM-1, PDGF-BB, and VEGF-A itself in NSCLC cells (42). They hypothesized that the VEGF-A feed-forward loop can induce the formation of a network of multiple growth factors sustaining angiogenesis in NSCLC cells (42). Such an angiogenic condition is expected to reduce sensitivity to anti-VEGF-A agents. In the present study, baseline serum concentrations of 12 proteins out of 25 analyzed angiogenesis-related ligands and receptors were significantly higher (FDR <0.05) in patients with serum follistatin above the cutpoint (≥490.5 pg/mL) than those in other patients (Figure S1). Follistatin and the 12 proteins detected include the same factors reported by Frezzetti et al., except leptin and VEGF-A (42). Although the participation of the VEGF-A feed-forward loop is not clear in the present study, high follistatin serum concentration may be a candidate tumor biomarker for the production of multiple angiogenic factors, and may be one explanation for the restricted efficacy of EB treatment in the patients with higher follistatin levels.
Approximately 40 patients with lower baseline serum levels of leptin were more sensitive to EB treatment than those with higher leptin levels. Leptin is a small adipokine (146 amino acids, 16 kDa) encoded by the obesity (ob) gene, which is primarily produced by fat cells (43). Serum leptin concentrations are highly correlated with body fat content and its production by adipocytes relates to nutritional status and acute phase reaction or chronic inflammation (44). In oncology, leptin has been investigated as an angiogenic factor that stimulates proliferation and reduces apoptosis in vascular ECs through leptin receptor expression (43). Leptin also causes the proliferation of various tumor cells such as BC and CRC, where leptin is known as an upstream regulator of VEGF-A and VEGFR2. Failure of initially promising anti-angiogenic therapies, and resistance to anti-angiogenic drugs mainly targeting VEGF-A/VEGFR2, including bevacizumab, are currently relevant problems (43). Compensatory and redundant effects of other angiogenic factors such as leptin are being investigated to clarify these failures.
Leptin levels are correlated with low fat mass or body mass index, and are thought to be inversely correlated with poor nutritional status. Patients with advanced NSCLC frequently suffer weight loss and malnutrition, where a low level of serum leptin is observed (44). Furthermore, patients with NSCLC and cachexia or anorexia are known to have significantly low serum leptin concentrations (44). The correlation with leptin levels and prognosis is unclear (45).
The relationship between serum leptin levels and local leptin levels produced in tumor tissue is not entirely known, but low serum leptin levels are expected to be caused by low levels of both local and systemic leptin production. In fact, the cutpoint of 1,662.4 pg/mL (approximately 1.7 ng/mL) evaluated in the current study was much lower than those considered in three representative studies in patients with NSCLC with comparatively large sample sizes: 9.3±10.8 ng/mL (n=101), 7.11±0.91 ng/mL (n=76) and 9.3±9.2 ng/mL (n=66) (44,46,47).
Patients with low leptin levels are thought to be sensitive to bevacizumab treatment, where leptin contributes little to local tumor angiogenesis, and they are anticipated to have anorexia or cachexia with a worse prognosis (43,44). Bevacizumab could inhibit tumor progression and induce apoptosis efficiently, which could lead to a reduction in the production of proinflammatory cytokines and/or growth factors in tumor cells, mediating cachexia and/or anorexia. These synergistic effects are speculated to improve EB treatment outcomes in patients with low levels of serum leptin.
In the current study, patients who are homozygous with minor allele VEGF-A rs3025039 SNP (T/T) and rs699947 (A/A) are suggested to be sensitive to EB treatment, as reported by other groups (19). However, we could not conclude that these SNPs are candidate predictors due to the small number of carriers with homozygous minor alleles. The minor allele (G) of SNP rs9582036 in VEGFR1 has been reported to be associated with improved EB treatment outcomes (OS and PFS) in patients with metastatic PAC (24). The marginally dominant effects of VEGFR1 rs9582036 on EB treatment outcomes were suggested in the present study but the effects were inconsistent between patients with G/G and G/T (Figure 5B). Low sensitivity to bevacizumab was reported in carriers of the VEGFR1 SNP rs7993418 C/C, but the study did not provide any information, since no patient had a C allele (24).
Since the present study was planned as an exploratory biomarker study, there are several limitations. First, there were no preplanned sample sizes or statistical significance levels for each biomarker analysis. Second, no external/internal validation studies were performed. Following the promising results of the JO25567 study, several phase III studies demonstrated consistent PFS improvement with EGFR-TKI treatment in combination with VEGF signaling inhibitors compared with EGFR-TKI monotherapy (48-52). We expect our results will provide a basis for the validation studies and contribute toward understanding the therapeutic positioning of the combination therapy among various solutions to improve EGFR-TKI outcomes for patients with EGFR mutation-positive NSCLC. Further biomarker studies are eagerly awaited.
Conclusions
In conclusion, our analysis of potential predictive biomarkers for favorable EB treatment outcomes in patients with EGFR-positive NSCLC reveal that (I) low levels of pVEGFA are associated with better EB treatment outcomes; (II) low leptin levels in baseline serum may be a potential biomarker for sensitivity to EB treatment; (III) high serum follistatin is a biomarker candidate for resistance to EB treatment and production of multiple growth factors; and (IV) a clinically meaningful predictive capacity could not be concluded from the angiogenesis-related gene polymorphisms analyzed and mRNAs/proteins in tumor tissue due to a shortage of samples from the patient population of interest.
Acknowledgments
We thank the patients, their families, and all of the investigators who participated in the study. The JO25567 study was sponsored by Chugai Pharmaceutical Co., Ltd. Third party writing assistance under the direction of the authors for this manuscript was provided by Shin-ichi Kaiho, freelance medical writer, funded by Chugai Pharmaceutical Co., Ltd., and Phoebe Liddell, MSc, and Fiona Duthie, PhD, of Ashfield MedComms, an Inizio Company, funded by Chugai Pharmaceutical Co., Ltd. Parts of the present study were presented at American Society of Clinical Oncology Annual Meeting, Chicago, IL, USA, May 30 to June 3, 2014; and the 16th World Conference on Lung Cancer, Denver, Colorado, USA, September 6 to 9, 2015.
Funding: This work was supported by Chugai Pharmaceutical Co., Ltd. No grant number is applicable.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-632/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-632/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-632/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-632/coif). TH serves as an unpaid editorial board member of Translational Lung Cancer Research from January 2022 to December 2023. MN reports receiving grants and personal fees from AstraZeneca, Amgen, MSD, Taiho Pharmaceutical, Takeda Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, Novartis, Pfizer, Bristol-Myers Squibb, and Merck Biopharma; personal fees from Ono Pharmaceutical, Boehringer Ingelheim, and Janssen Pharmaceutical; and grants from Daiichi-Sankyo. SA reports grants from AstraZeneca, Eli Lilly, Ono, Taiho Pharmaceutical, Boehringer Ingelheim, Pfizer, Bristol-Myers Squibb, MSD, Chugai Pharmaceutical, Merck, and F. Hoffmann-La Roche; and honoraria from Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, Sawai Pharmaceutical, Novartis Pharma, AstraZeneca, Ono, Boehringer Ingelheim, Pfizer, Bristol-Myers Squibb, Hisamitsu, MSD, Kyowa Hakko Kirin, Merck, and Thermo Fisher Scientific. KG reports receiving research grants from Amgen Astellas BioPharma, Amgen, AstraZeneca, Bayer Yakuhin, Ltd., Bristol-Myers Squibb K.K., Boehringer Ingelheim Japan, Inc., Blueprint Medicines Corporation, Daiichi-Sankyo Co., Ltd., Eisai Co., Ltd., Eli Lilly Japan K.K., Haihe Biopharma Co., Ltd., Ignyta, Inc., Janssen Pharmaceutical K.K., Kissei Pharmaceutical Co., Ltd., Kyowa Kirin Co., Ltd., Life Technologies Japan Ltd., Loxo Oncology, Inc., Medical & Biological Laboratories Co., Ltd., Merck Biopharma Co., Ltd., Merus N.V., MSD K.K., NEC Corporation, Novartis Pharma K.K., Ono Pharmaceutical Co., Ltd., Pfizer Japan Inc., Sumitomo Dainippon Pharma Co., Ltd., Spectrum Pharmaceuticals, Inc., Sysmex Corporation, Taiho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., and Turning Point Therapeutics, Inc.; honoraria from Amgen Inc., AstraZeneca K.K., Bayer HealthCare Pharmaceuticals Inc., Boehringer Ingelheim Japan, Inc., Bristol-Myers Squibb K.K., Daiichi Sankyo Co., Ltd., Eisai Co., Ltd., Eli Lilly Japan K.K., Guardant Health Inc., Janssen Pharmaceutical K.K., Kyowa Kirin Co., Ltd., Life Technologies Japan Ltd., Medpace Japan K.K., Merck Biopharma Co., Ltd., MSD K.K., Novartis Pharma K.K., Ono Pharmaceutical Co., Ltd., Otsuka Pharmaceutical Co., Ltd., Pfizer Japan Inc., Taiho Pharmaceutical Co., Ltd., and Takeda Pharmaceutical Co., Ltd. YH reports receiving payment/honoraria from Chugai Pharmaceutical, AstraZeneca, Eli Lilly Japan, Taiho Pharmaceutical, Bristol-Myers Squibb, Kyowa Kirin, and Ono Pharmaceutical. TS reports receiving institutional funding from AbbVie, Chugai Pharmaceutical, Daiichi Sankyo, Eli Lilly Japan, Kissei Pharmaceutical, MSD, Novartis Pharma, Pfizer Japan, Takeda Pharmaceutical; honoraria from AstraZeneca, Bristol-Myers Squibb, Chugai Pharmaceutical, Covidien Japan, Daiichi Sankyo, Eli Lilly Japan, Kyowa Hakko Kirin, MSD, Mochida Pharmaceutical, Nippon Boehringer Ingelheim, Novartis Pharma, Ono Pharmaceutical, Pfizer Japan, Taiho Pharmaceutical, Takeda Pharmaceutical, and Towa Pharmaceutical; and is an employee of Precision Medicine Asia. TH reports receiving research grants from Chugai Pharmaceutical, AstraZeneca, Nippon Boehringer Ingelheim, Pfizer, Eli Lilly, Takeda Bio, Novartis Pharma, Ono Pharmaceutical, Daiichi-Sankyo, Merck Serono, Dainippon Sumitomo Pharma, Bristol-Myers Squibb, and Eisai. Kazuhiko Nakagawa reports receiving honoraria from Chugai Pharmaceutical, AstraZeneca, Nippon Boehringer Ingelheim, and Eli Lilly. HY reports research grants from Chugai Pharmaceutical, Boehringer Ingelheim, and Nippon Kayaku; consulting fees from Delta Fly Pharma; honoraria from Eli Lilly, Chugai Pharmaceutical, Boehringer Ingelheim, Delta Fly Pharma, AstraZeneca, BMS, MSD, Ono Pharmaceutical, Merck Biopharma, Nippon Kayaku, Taiho Pharmaceutical, Otsuka Pharmaceutical, Takeda Pharmaceutical, Novartis Pharmaceutical, Amgen, Nipro Pharma, Kyowa Kirin, and Pfizer. NN reports receiving honoraria from Chugai Pharmaceutical, Pfizer Japan, Taiho Pharmaceutical, Daiichi Sankyo, Eli Lilly Japan, AstraZeneca, and Nippon Boehringer Ingelheim. MM reports receiving honoraria from Chugai Pharmaceutical and AstraZeneca; research grants and honoraria from Nippon Boehringer Ingelheim. IO reports receiving payment/honoraria from Chugai Pharmaceutical. Noboru Yamamoto reports receiving research grants from Chugai Pharmaceutical, Taiho Pharmaceutical, Eisai, Eli Lilly, Quintiles, Astellas Pharma, Bristol-Myers Squibb, Novartis, Daiichi-Sankyo, Pfizer, Boehringer Ingelheim, Kyowa-Hakko Kirin, Bayer, Ono Pharmaceutical Co., Ltd., Takeda, Janssen Pharma, MSD, Merck, GSK, Sumitomo Dainippon, Chiome Bioscience Inc, Otsuka, Carna Bioscienes, Genmab, and Shionogi; honoraria from Ono Pharmaceutical, Chugai Pharmaceutical, AstraZeneca, Pfizer, Eli Lilly, Bristol-Myers Squibb, Sysmex, and Eisai; and consulting fees from Eisai, Otsuka, Takeda, Boehringer Ingelheim, and Cimic. YI and KT are employees of Chugai Pharmaceutical. MF reports receiving honoraria from Eisai and Chugai Pharmaceutical. Nobuyuki Yamamoto reports receiving grants from MSD K.K., AstraZeneca, Ono Pharmaceutical Co., Ltd., Daiichi-Sankyo Co., Ltd., Taiho Pharmaceutical., Takeda, Chugai Pharmaceutical Co., Ltd., Pfizer, Amgen, Janssen Pharmaceutical K.K., Toppan Printing, Terumo, and Boehringer Ingelheim; consulting fees from AstraZeneca, Daiichi-Sankyo, Taiho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Eli Lilly Japan K.K., Boehringer Ingelheim, Novartis, Bristol-Myers Squibb, Nippon Kayaku, Life Technologies Japan, Ltd., Amgen, Guardant Health Japan, and Janssen Pharmaceutical K.K.; honoraria from MSD, AstraZeneca, Ono Pharmaceutical, Thermo Fisher Scientific, Daiichi-Sankyo, Taiho Pharmaceutical, Takeda, Chugai Pharmaceutical, Eli Lilly Japan K.K., Boehringer Ingelheim, Novartis, Pfizer, Bristol-Myers Squibb, Nippon Kayaku, GSK K.K., Sanofi K.K., Hisamitsu Pharmaceutical, and Merck Biopharma; and participated on a data safety monitoring board/advisory board for MSD K.K., AstraZeneca, Ono Pharmaceutical, Takeda Pharmaceutical, Chugai Pharmaceutical, Eli Lilly Japan K.K., Boehringer Ingelheim, and Pfizer. Kazuto Nishio reports receiving grants from Nippon Boehringer Ingelheim, West Japan Oncology Group, Thoracic Oncology Research Group, North East Japan Study Group, Clinical Research Support Center Kyushu, Nichirei Biosciences Inc., and Eli Lilly Japan; consulting fees from SymBio Pharmaceuticals, Solasia Pharma, Eli Lilly Japan, and Otsuka Pharmaceutical; honoraria for lectures from Boehringer Ingelheim Japan, AstraZeneca, Chugai, Novartis Pharma, Eisai, MSD, Bristol-Myers Squibb, Ono Pharmaceutical, Pfizer, Sanofi, Guardant Health, Eli Lilly Japan, Amgen, Merck Biopharma, Roche Diagnostics, Yakult Honsha, Takeda Pharmaceutical, Fujirebio, and Janssen Pharmaceutical. All authors report receiving funding from Chugai Pharmaceutical Co., Ltd. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and Good Clinical Practice guidelines. The study protocol was reviewed and approved by the institutional review boards of the participating institutions, and written informed consent was obtained from all patients. The study was registered with the Japan Pharmaceutical Information Center (No. JapicCTI-111390).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Graham RP, Treece AL, Lindeman NI, et al. Worldwide Frequency of Commonly Detected EGFR Mutations. Arch Pathol Lab Med 2018;142:163-7. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med 2006;355:2542-50. [Crossref] [PubMed]
- Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov 2016;15:385-403. [Crossref] [PubMed]
- Ferrara N, Hillan KJ, Gerber HP, et al. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov 2004;3:391-400. [Crossref] [PubMed]
- Le X, Nilsson M, Goldman J, et al. Dual EGFR-VEGF Pathway Inhibition: A Promising Strategy for Patients With EGFR-Mutant NSCLC. J Thorac Oncol 2021;16:205-15. [Crossref] [PubMed]
- Naumov GN, Nilsson MB, Cascone T, et al. Combined vascular endothelial growth factor receptor and epidermal growth factor receptor (EGFR) blockade inhibits tumor growth in xenograft models of EGFR inhibitor resistance. Clin Cancer Res 2009;15:3484-94. [Crossref] [PubMed]
- Furugaki K, Fukumura J, Iwai T, et al. Impact of bevacizumab in combination with erlotinib on EGFR-mutated non-small cell lung cancer xenograft models with T790M mutation or MET amplification. Int J Cancer 2016;138:1024-32. [Crossref] [PubMed]
- Seto T, Kato T, Nishio M, et al. Erlotinib alone or with bevacizumab as first-line therapy in patients with advanced non-squamous non-small-cell lung cancer harbouring EGFR mutations (JO25567): an open-label, randomised, multicentre, phase 2 study. Lancet Oncol 2014;15:1236-44. [Crossref] [PubMed]
- Van Cutsem E, de Haas S, Kang YK, et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a biomarker evaluation from the AVAGAST randomized phase III trial. J Clin Oncol 2012;30:2119-27. [Crossref] [PubMed]
- Miles DW, de Haas SL, Dirix LY, et al. Biomarker results from the AVADO phase 3 trial of first-line bevacizumab plus docetaxel for HER2-negative metastatic breast cancer. Br J Cancer 2013;108:1052-60. [Crossref] [PubMed]
- Van Cutsem E, Jayson G, Dive C, et al. Analysis of blood plasma factors in the AVITA phase III randomized study of bevacizumab (bev) with gemcitabine-erlotinib (GE) in patients (pts) with metastatic pancreatic cancer (mPC). Eur J Cancer 2011;47:S95-6. [Crossref]
- Bais C, Rabe C, Wild N, et al. Comprehensive reassessment of plasma VEGFA (pVEGFA) as a candidate predictive biomarker for bevacizumab (Bv) in 13 pivotal trials (seven indications). J Clin Oncol 2014;32:abstr 3040.
- Lambrechts D, Lenz HJ, de Haas S, et al. Markers of response for the antiangiogenic agent bevacizumab. J Clin Oncol 2013;31:1219-30. [Crossref] [PubMed]
- Garcia V, Garcia JM, Silva J, et al. Levels of VEGF-A mRNA in plasma from patients with colorectal carcinoma as possible surrogate marker of angiogenesis. J Cancer Res Clin Oncol 2008;134:1165-71. [Crossref] [PubMed]
- Bates DO, Catalano PJ, Symonds KE, et al. Association between VEGF splice isoforms and progression-free survival in metastatic colorectal cancer patients treated with bevacizumab. Clin Cancer Res 2012;18:6384-91. [Crossref] [PubMed]
- Pentheroudakis G, Kotoula V, Kouvatseas G, et al. Association of VEGF-A splice variant mRNA expression with outcome in bevacizumab-treated patients with metastatic breast cancer. Clin Breast Cancer 2014;14:330-8. [Crossref] [PubMed]
- Jubb AM, Miller KD, Rugo HS, et al. Impact of exploratory biomarkers on the treatment effect of bevacizumab in metastatic breast cancer. Clin Cancer Res 2011;17:372-81. [Crossref] [PubMed]
- Lambrechts D, Claes B, Delmar P, et al. VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol 2012;13:724-33. [Crossref] [PubMed]
- Krintel SB, Grunert VP, Hetland ML, et al. The frequency of anti-infliximab antibodies in patients with rheumatoid arthritis treated in routine care and the associations with adverse drug reactions and treatment failure. Rheumatology (Oxford) 2013;52:1245-53. [Crossref] [PubMed]
- Bonetti M, Gelber RD. A graphical method to assess treatment-covariate interactions using the Cox model on subsets of the data. Stat Med 2000;19:2595-609. [Crossref] [PubMed]
- Royston P, Sauerbrei W. A new approach to modelling interactions between treatment and continuous covariates in clinical trials by using fractional polynomials. Stat Med 2004;23:2509-25. [Crossref] [PubMed]
- Altman DG, McShane LM, Sauerbrei W, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med 2012;9:e1001216. [Crossref] [PubMed]
- Jiang W, Freidlin B, Simon R. Biomarker-adaptive threshold design: a procedure for evaluating treatment with possible biomarker-defined subset effect. J Natl Cancer Inst 2007;99:1036-43. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57:289-300. [Crossref]
- Miles D, Cameron D, Bondarenko I, et al. Bevacizumab plus paclitaxel versus placebo plus paclitaxel as first-line therapy for HER2-negative metastatic breast cancer (MERiDiAN): A double-blind placebo-controlled randomised phase III trial with prospective biomarker evaluation. Eur J Cancer 2017;70:146-55. [Crossref] [PubMed]
- Bai L, Wang F, Zhang DS, et al. A plasma cytokine and angiogenic factor (CAF) analysis for selection of bevacizumab therapy in patients with metastatic colorectal cancer. Sci Rep 2015;5:17717. [Crossref] [PubMed]
- Sepporta MV, Tumminello FM, Flandina C, et al. Follistatin as potential therapeutic target in prostate cancer. Target Oncol 2013;8:215-23. [Crossref] [PubMed]
- Chen F, Ren P, Feng Y, et al. Follistatin is a novel biomarker for lung adenocarcinoma in humans. PLoS One 2014;9:e111398. [Crossref] [PubMed]
- Gökmen-Polar Y, Goswami CP, Toroni RA, et al. Gene Expression Analysis Reveals Distinct Pathways of Resistance to Bevacizumab in Xenograft Models of Human ER-Positive Breast Cancer. J Cancer 2014;5:633-45. [Crossref] [PubMed]
- Krneta J, Kroll J, Alves F, et al. Dissociation of angiogenesis and tumorigenesis in follistatin- and activin-expressing tumors. Cancer Res 2006;66:5686-95. [Crossref] [PubMed]
- Blank C, Mackensen A. Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: an update on implications for chronic infections and tumor evasion. Cancer Immunol Immunother 2007;56:739-45. [Crossref] [PubMed]
- Maeshima K, Maeshima A, Hayashi Y, et al. Crucial role of activin a in tubulogenesis of endothelial cells induced by vascular endothelial growth factor. Endocrinology 2004;145:3739-45. [Crossref] [PubMed]
- Ries A, Schelch K, Falch D, et al. Activin A: an emerging target for improving cancer treatment? Expert Opin Ther Targets 2020;24:985-96. [Crossref] [PubMed]
- Betsholtz C, Bergh J, Bywater M, et al. Expression of multiple growth factors in a human lung cancer cell line. Int J Cancer 1987;39:502-7. [Crossref] [PubMed]
- Occleston NL, Walker C. Production of multiple growth factors by a human non-small cell lung carcinoma cell line. Cancer Lett 1993;71:203-10. [Crossref] [PubMed]
- Frezzetti D, Gallo M, Roma C, et al. Vascular Endothelial Growth Factor A Regulates the Secretion of Different Angiogenic Factors in Lung Cancer Cells. J Cell Physiol 2016;231:1514-21. [Crossref] [PubMed]
- Gonzalez-Perez RR, Lanier V, Newman G. Leptin's Pro-Angiogenic Signature in Breast Cancer. Cancers (Basel) 2013;5:1140-62. [Crossref] [PubMed]
- Alemán MR, Santolaria F, Batista N, et al. Leptin role in advanced lung cancer. A mediator of the acute phase response or a marker of the status of nutrition? Cytokine 2002;19:21-6. [Crossref] [PubMed]
- Anar C, Deniz D, Erol S, et al. Are serum leptin levels a prognostic factor in advanced lung cancer? Bratisl Lek Listy 2017;118:13-6. [Crossref] [PubMed]
- Karapanagiotou EM, Tsochatzis EA, Dilana KD, et al. The significance of leptin, adiponectin, and resistin serum levels in non-small cell lung cancer (NSCLC). Lung Cancer 2008;61:391-7. [Crossref] [PubMed]
- Terzidis A, Sergentanis TN, Antonopoulos G, et al. Elevated serum leptin levels: a risk factor for non-small-cell lung cancer? Oncology 2009;76:19-25. [Crossref] [PubMed]
- Saito H, Fukuhara T, Furuya N, et al. Erlotinib plus bevacizumab versus erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomised, multicentre, phase 3 trial. Lancet Oncol 2019;20:625-35. [Crossref] [PubMed]
- Zhou Q, Xu CR, Cheng Y, et al. Bevacizumab plus erlotinib in Chinese patients with untreated, EGFR-mutated, advanced NSCLC (ARTEMIS-CTONG1509): A multicenter phase 3 study. Cancer Cell 2021;39:1279-91.e3. [Crossref] [PubMed]
- Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:1655-69. [Crossref] [PubMed]
- Zhao H, Yao W, Min X, et al. Apatinib Plus Gefitinib as First-Line Treatment in Advanced EGFR-Mutant NSCLC: The Phase III ACTIVE Study (CTONG1706). J Thorac Oncol 2021;16:1533-46. [Crossref] [PubMed]
- Piccirillo MC, Bonanno L, Garassino MC, et al. Addition of Bevacizumab to Erlotinib as First-Line Treatment of Patients With EGFR-Mutated Advanced Nonsquamous NSCLC: The BEVERLY Multicenter Randomized Phase 3 Trial. J Thorac Oncol 2022;17:1086-97. [Crossref] [PubMed]


