
A rationale for the poor response to alectinib in a patient with adenocarcinoma of the lung harbouring a STRN-ALK fusion by artificial intelligence and molecular modelling: a case report
Highlight box
Key findings
• A patient with a stage IV adenocarcinoma of the lung with STRN-ALK fusion and STK11-LKB1 c.290+1G>A mutation underwent alectinib treatment with partial response but developed progression after six months.
• The change in therapy to lorlatinib was ineffective and the patient developed an unstoppable thoracic and bone progression.
• Retrospective evaluation with Artificial Intelligence and Molecular Modelling tools showed that the fusion distorted the drug binding pocket.
What is known and what is new?
• Different clinical trials showed the efficacy of alectinib and lorlatinib in patients with adenocarcinoma of the lung harbouring ALK fusions.
• We report a patient with a poor response to these drugs proposing new tools for a mechanistic explanation of the clinical findings.
What is the implication, and what should change now
• Molecular modelling can offer potentially important predictive informations in patient with rare ALK fusions.
• Functional assays and prospective studies are warranted to demonstrate the predictive robustness of these tools.
Introduction
Anaplastic lymphoma kinase (ALK) gene rearrangements (fusions) are oncogenic drivers detected in many solid and hematologic malignancies, including histiocytosis, leiomyosarcoma, lymphoma, myeloma, and lung, colorectal, neuroendocrine, ovarian, pancreatic, renal and thyroid cancers. In non-small cell lung cancer (NSCLC), mainly in adenocarcinoma, ALK rearrangement occurs in 4–7% of cases (1). This alteration confers sensitivity to ALK inhibition using tyrosine kinase inhibitors (TKI). Crizotinib, a first-in-class ALK/ROS1/MET inhibitor has shown advantage against chemotherapy in two randomized phase III trials (2,3). Since then, seven ALK-TKI (crizotinib, ceritinib, alectinib, brigatinib, ensartinib, lorlatinib, iruplinakib) have been approved in different parts of the world for the first-line treatment of advanced ALK-positive NSCLC (4). A recent study comparing lorlatinib versus crizotinib in patients with advanced ALK-positive NSCLCs showed that after 5 years of follow-up, median progression-free survival (PFS) was still to be achieved in the lorlatinib group, “corresponding to the longest PFS ever reported with any single-agent molecular targeted treatment in advanced NSCLC and across all metastatic solid tumors” (5). In spite of these advances, the management of these tumors remains challenging due to the presence of the rare primary (intrinsic) or the more common acquired mechanisms of resistance to TKIs (6). Echinoderm-microtubule-associated protein like 4 (EML4) is the most common fusion partner of ALK, but the widespread use of next-generation sequencing (NGS) assays to study the molecular profile of NSCLCs, can identify rare fusion partners of ALK. Therapy decisions are made without considering which fusion partner is present and its potential oncogenic properties. However, clinical and experimental studies have shown that the 5' partner of kinase fusion variants could have a biological role in the response to targeted therapies (7,8). Here we describe a case of lung adenocarcinoma with a rare striatin (STRN)-ALK fusion, a calmodulin-binding scaffolding protein regulating multiple signaling pathways and cytoskeleton remodelling, and discuss the possibility of using artificial intelligence (AI) tools and molecular modelling to predict response to ALK TKI. We present this article in accordance with the CARE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-667/rc).
Case presentation
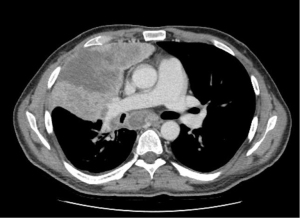
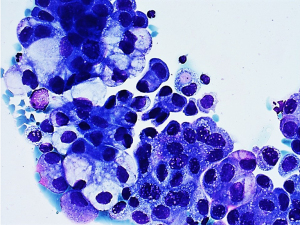
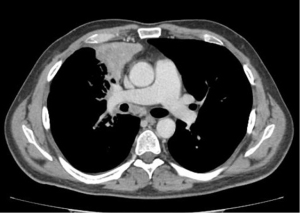
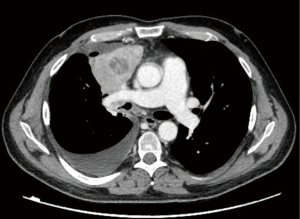
A 45-year-old male with no history of smoking suddenly developed severe back pain and widespread cutaneous tense blisters. After an ineffective symptomatic therapy, a computed tomography (CT)-scan of the chest revealed a voluminous mass of the right upper lobe associated with homolateral and mediastinal lymphadenopathies (Figure 1). Fluorodeoxyglucose (FDG)-positron emission tomography confirmed the pulmonary and lymph-nodal involvement and showed the presence of bone metastases in the second lumbar vertebra and the fifth right rib. After two months from the beginning of symptoms, the patient was admitted at the European Institute of Oncology with a clinical diagnosis of advanced (stage IV) lung tumor associated with a paraneoplastic pemphigoid. The obtained medical, family and genetic information were not relevant. He underwent endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) of a 4R lymph node. May-Grunwald stained smears (Figure 2) and hematoxylin & eosin-stained sections from cell-block showed aggregates of malignant epithelial cells with foamy cytoplasm. Immunocytochemistry revealed nuclear positivity for TTF1 and intense cytoplasmatic staining for cytokeratin 7 and ALK (clone D5F3, Ventana, Tucson, AZ, USA). No thyroglobulin expression was found. Morphology and phenotype were consistent with a lung adenocarcinoma. DNA and RNA were then extracted from the cytological sample and subjected to NGS analysis using the Oncomine Comprehensive Assay v3 (Thermofisher Scientific, Waltham, MA, USA). A fusion between STRN (locus 2p22) and ALK gene (locus 2p23) was identified together with a pathogenetic single nucleotide variant of STK11-LKB1 gene (c.290+1G>A) affecting a donor splice site in intron 1. It displayed a low allelic frequency (6%) suggesting a subclonal event. No other genetic aberrations were found. The patient received alectinib at the standard dose of 600 mg twice a day. A radiological partial response was observed after three months of treatment (Figure 3). Clinical regression of the paraneoplastic pemphigus occurred after two weeks of treatment with alectinib.
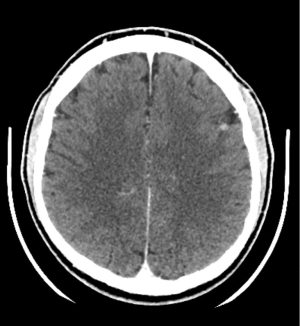
After six months of treatment, a CT scan showed disease progression with development of brain metastases (Figure 4); in the same time there was a widespread recurrence of the paraneoplastic pemphigus. In Europe Lorlatinib was approved as monotherapy for the treatment of adult patients with ALK-positive advanced NSCLC whose disease has progressed after: alectinib or ceritinib as the first ALK TKI therapy (https://www.ema.europa.eu/en/documents/product-information/lorviqua-epar-product-information_en.pdf). Second-line treatment with lorlatinib was tried but the patient had unstoppable thoracic (Figure 5) and bone progression within two months, leading to death. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal. The patient specific information were de-identified.
Discussion
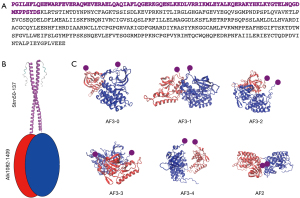
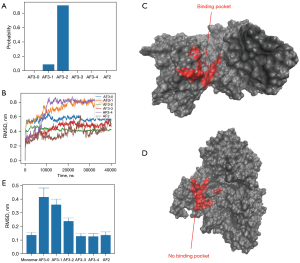
Since the first identification of the most common ALK fusion gene (EML4 ALK) in NSCLC (9), more than 30 5’-partner genes of ALK have been reported on. These partners play a critical role in the biological functions of the chimeric protein (7,8,10). The response rate of rare ALK fusions to different ALK-TKIs is based on individual case reports. Recently, Song et al. reported a case of NSCLC with STRN-ALK fusion, with additional eleven cases with the same fusion being found by literature review (11). Differences in treatment (chemotherapy, crizotinib, alectinib, lorlatinib, ensartinib), lack of follow up for some patients, and the presence of co-mutations, mainly p53, led to conflicting results, which Li et al. correctly defined “mixed responses” (12). In our patient, we observed an initial clinical benefit from first-line treatment with alectinib but disease progressed rapidly despite prompt switching to lorlatinib. Therefore, we tried to get more information about treatment options offered to our patient using molecular modelling based on an AI tool. Briefly, we investigated a computational molecular model of the protein translated from the translocated gene to suggest a mechanistic explanation for the clinical findings. The protein consisted of residues 1–137 of STRN and 1082–1620 of ALK. AlphaFold3 (13) predicted that residues 1–49 and 121–137 of STRN and 1410–1620 of ALK were intrinsically disordered. We modelled (Figure 6A) the complex made by STRN50–137 and ALK1082–1409. AlphaFold3 predicted a pair of two identical chains of STRN50-137 to form a dimeric coiled-coil structure (Figure 6B) with high confidence (ipTM=0.67). An estimate of the binding free energy of two STRN50–137 fragments made with FoldX (14) also indicated a strong propensity to dimerize (DG=−29.4 kcal/mol). Our working hypothesis was then that the dimerization of STRN stabilized a dimer of ALK, which was tethered to it by the translocation. In the absence of biochemical data, we have used model calculations to predict the most likely structure of the dimer of ALK1082–1409. We used the five best dimeric models predicted by AlphaFold3 and one predicted by AlphaFold2 as a starting point (Figure 6C). The confidence level of all these models is very low (ipTM <0.2). This is not unexpected since they are not dimers that could have been formed spontaneously in the absence of the tethering to STRN. We performed a 40 ns of molecular dynamics simulation of each of these initial structures [using Gromacs (15) with the Amber03 force field and Tip3p water at 300K (16)]. During the simulation time, the six models underwent some conformational rearrangement to varying degrees (Figure 6C) but remained dimeric. The average free energy of dissociation of the dimer was estimated with FoldX and used to calculate the relative probability of the models according to the Boltzmann distribution. The most probable models was AF3-2 and, to a lesser extent, AF3-1 (Figure 7A). This result was justified by the higher protection of the hydrophobic residues offered by dimerization in model AF3-2 than in the other models (DGhydrophobic=−45.5 kcal/mol, e.g., compared to (DGhydrophobic=−16.7 kcal/mol of model AF3-4). We then examined the deformation during the simulation (Figure 7B) of the single binding pocket to alectinib, lorlatinib and crizotinib, as obtained from the crystal structures of the native protein bound to these ligands (PDB codes 3a0x, 5a9u and 2xp2, respectively). Even in the apo form, the monomeric protein displayed a pocket that was stably open in our simulations (Figure 7C). Upon dimerization into the AF3-2 structure, the pocket closed up and thus becomes inaccessible to the ligands (Figure 7D). The deformation of the pocket in both the dimeric models AF3-1 and AF3-2 was consistently larger than the spontaneous thermal deformation of the monomeric model (Figure 7E).
In summary, model calculations suggested that the effect of the translocation was to induce the dimerization of ALK into a complex that closed the binding pocket, which is the same for alectinib, lorlatinib and crizotinib. The distortion of the binding pocket observed in the simulations also provides a rationale to explain the different variations of efficacy of alectinib, lorlatinib and crizotinib caused by the translocation. Indeed, all of them target the same pocket (cf. crystallographic structures 3AOX, 5A9U and 2XP2 in the Protein Data Bank).
These observations raised the question as to whether crizotinib might have given better response to our patient.
Crizotinib has a conformational flexibility much larger than alectinib and lorlatinib due to the presence of 5 routable bonds, instead of 3 and 0, respectively (17). Consequently, crizotinib has a larger IC50 against the wild-type protein (60.6 nM, compared to 3.5 and 1.3 nM) (18,19), respectively, due to the higher entropic cost of binding a rigid target. However, it is conceivable that its very flexibility could have allowed it to adapt its structure to the distorted and fluctuating pocket better than the more rigid molecules alectinib and lorlatinib.
Interestingly in NSCLC, STRN-ALK fusion has previously been reported to be responsive to crizotinib (20) and not responsive to alectinib (21).
We also detected a co-existing mutation of STK11/LKB1. These alterations have been associated with poor prognosis in several therapeutic classes of NSCLC (22,23).
Conclusions
In conclusion, our observations suggest that molecular modelling based on AI tools may offer potential predictive information in fusions with rare partner genes. Further retrospective and prospective studies are warranted to demonstrate the predictive robustness of these tools. We will develop a project to create a comprehensive model of therapy resistance of rare ALK fusions. To achieve this, we will combine AlphaFold protein folding predictions with molecular dynamics (MD) simulations to generate a dynamic computational model of the protein encoded by the translocated gene. This approach will allow us to explore the resulting alterations in protein conformation and assess the consequent changes in drug binding affinity. Following its development, the model will be assessed using a real-world cohort of patients affected by NSCLC harboring ALK fusions. Clinicogenomic data will be retrieved retrospectively, and clinical outcomes measured.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-667/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-667/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-667/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chia PL, Mitchell P, Dobrovic A, et al. Prevalence and natural history of ALK positive non-small-cell lung cancer and the clinical impact of targeted therapy with ALK inhibitors. Clin Epidemiol 2014;6:423-32. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Ou SI, Lee ATM, Nagasaka M. From preclinical efficacy to 2022 (36.7 months median follow -up) updated CROWN trial, lorlatinib is the preferred 1st-line treatment of advanced ALK+ NSCLC. Crit Rev Oncol Hematol 2023;187:104019. [Crossref] [PubMed]
- Solomon BJ, Liu G, Felip E, et al. Lorlatinib Versus Crizotinib in Patients With Advanced ALK-Positive Non-Small Cell Lung Cancer: 5-Year Outcomes From the Phase III CROWN Study. J Clin Oncol 2024;42:3400-9. [Crossref] [PubMed]
- Schneider JL, Lin JJ, Shaw AT. ALK-positive lung cancer: a moving target. Nat Cancer 2023;4:330-43. [Crossref] [PubMed]
- Childress MA, Himmelberg SM, Chen H, et al. ALK Fusion Partners Impact Response to ALK Inhibition: Differential Effects on Sensitivity, Cellular Phenotypes, and Biochemical Properties. Mol Cancer Res 2018;16:1724-36. [Crossref] [PubMed]
- Yoshida T, Oya Y, Tanaka K, et al. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383-9. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Song GQ, Li YZ, Kong W, et al. Case Report: A rare case of non-small cell lung cancer with STRN-ALK fusion in a patient in very poor condition treated with first-line ensartinib. Front Oncol 2023;13:1235679. [Crossref] [PubMed]
- Li M, An Z, Tang Q, et al. Mixed responses to first-line alectinib in non-small cell lung cancer patients with rare ALK gene fusions: A case series and literature review. J Cell Mol Med 2021;25:9476-81. [Crossref] [PubMed]
- Delgado J, Radusky LG, Cianferoni D, et al. FoldX 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics 2019;35:4168-9. [Crossref] [PubMed]
- Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024;630:493-500. [Crossref] [PubMed]
- Abraham MJ, Murtola T, Schulz R, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015;1–2:19-25. [Crossref]
- Wang J, Wolf RM, Caldwell JW, et al. Development and testing of a general amber force field. J Comput Chem 2004;25:1157-74. [Crossref] [PubMed]
- Kim S, Chen J, Cheng T, et al. PubChem 2023 update. Nucleic Acids Res 2023;51:D1373-80. [Crossref] [PubMed]
- Amin AD, Li L, Rajan SS, et al. TKI sensitivity patterns of novel kinase-domain mutations suggest therapeutic opportunities for patients with resistant ALK+ tumors. Oncotarget 2016;7:23715-29. [Crossref] [PubMed]
- Geng K, Xia Z, Ji Y, et al. Discovery of 2,4-diarylaminopyrimidines bearing a resorcinol motif as novel ALK inhibitors to overcome the G1202R resistant mutation. Eur J Med Chem 2018;144:386-97. [Crossref] [PubMed]
- Yang Y, Qin SK, Zhu J, et al. A Rare STRN-ALK Fusion in Lung Adenocarcinoma Identified Using Next-Generation Sequencing-Based Circulating Tumor DNA Profiling Exhibits Excellent Response to Crizotinib. Mayo Clin Proc Innov Qual Outcomes 2017;1:111-6. [Crossref] [PubMed]
- Nakanishi Y, Masuda S, Iida Y, et al. Case Report of Non-Small Cell Lung Cancer with STRN-ALK Translocation: A Nonresponder to Alectinib. J Thorac Oncol 2017;12:e202-4. [Crossref] [PubMed]
- Pécuchet N, Laurent-Puig P, Mansuet-Lupo A, et al. Different prognostic impact of STK11 mutations in non-squamous non-small-cell lung cancer. Oncotarget 2017;8:23831-40. [Crossref] [PubMed]
- Papillon-Cavanagh S, Doshi P, Dobrin R, et al. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open 2020;5:e000706. [Crossref] [PubMed]


