
Construction of a neutrophil extracellular trap formation-related gene model for predicting the survival of lung adenocarcinoma patients and their response to immunotherapy
Highlight box
Key findings
• Revealing the expression patterns of neutrophil extracellular traps formation-related genes (NFRGs) and construction a predictive model for lung adenocarcinoma (LUAD) by integrative analysis of single-cell and bulk transcriptomes.
What is known and what is new?
• Studies have shown that NETs play critical roles in tumor progression, metastasis and immunosuppression in the LUAD tumor microenvironment (TME).
• We constructed a prediction model for the survival, drug sensitivity, immune checkpoint blockade (ICB) response for LUAD patients based on four NFRGs, including CAT, CTSG, TLR2, ENO1.
What is the implication, and what should change now?
• This study highlights the value of NETs in stratifying LUAD patients and informing individualized management strategies.
• Efforts should be taken to adjust and explore the predictive value of our risk score for ICB response in multicenter and large-scale prospective cohorts.
Introduction
Lung cancer is highly aggressive and heterogeneous, with a distinct mutation load and histological status, and is the leading cause of cancer-related death (1,2). Lung adenocarcinoma (LUAD) is the most common type of lung cancer, accounting for 40% of all lung cancer types and approximately 55% of non-small cell lung cancers (NSCLCs). In addition to driving tumor progression and evolution, genetic heterogeneity helps shape the tumor microenvironment (TME). With advances in high-throughput sequencing technology, researchers have gained new insights into the composition and function of the TME in LUAD (3-6).
Solid tumors accumulate a complex set of innate and acquired immune inflammatory cells in the TME. It is generally believed that tumors are heavily infiltrated with neutrophils and that the prognosis is relatively poor. Neutrophils represent 50–70% of circulating leukocytes in humans and are considered the “soldiers” of the innate immune system with a restricted set of pro-inflammatory functions. However, in the TME, high infiltration of tumor-associated neutrophils (TANs) has been associated with poor prognosis (7).
Neutrophil extracellular traps (NETs) are networks of DNA histones and proteins released by activated neutrophils. Despite helping to immobilize and eliminate pathogens (8-10), an increasing number of studies have demonstrated that NETs play a significant role in tumor progression and metastasis (11-16). NETs are quantified by the area of colocalization of histone markers or extracellular DNA with neutrophil granular or cytoplasmic components (17), which can help to predict clinical stage and prognosis (18-22). The immunosuppressive role of NETs in the TME mainly involves triggering the exhaustion and dysfunction of CD8+ T cells (23-25), regulating the balance between Th1 and Th2 CD4+ T cells and impairing the antitumor effects of natural killer (NK) cells (26-30).
The capability of NET formation-related genes (NFRGs) to evaluate LUAD patient survival and response to immunotherapy is not clear. In this study, we constructed an NFRGs-based prognostic signature for LUAD patients to predict survival and immunotherapy response. First, we explored the distinct composition of the TME and cell-cell interactions in LUAD patients at single-cell resolution and noted a substantial increase in neutrophils in the TME. We then comprehensively assessed the expression of 24 neutrophil trap formation-related genes at both the single-cell level and bulk RNA level. Next, we constructed and validated a 4-NFRG-based prognostic model for LUAD patients based on The Cancer Genome Atlas (TCGA) Program, Gene Expression Omnibus (GEO) and real-world clinical cohort data. The potential of our risk score to stratify LUAD patients with different clinical information was evaluated. Subsequently, we developed a nomogram to quantify survival probability combined with the risk score and other clinical characteristics. Furthermore, the predictive value of this nomogram for prognosis, immunotherapy efficacy and chemotherapy sensitivity were assessed and explored based on distinct datasets. We present this article in accordance with the TRIPOD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-463/rc).
Methods
Single‑cell RNA sequence analysis
The “NormalizeData” function was used to normalize raw counts, and the “FindVariableFeatures” function was utilized to select highly variable genes in the R package “Seurat” 43 (V4.0.3) to address the batch effect and tissue specificity of the single-cell RNA-seq data. Principal component analysis (PCA) and uniform manifold approximation and projection for dimension reduction (UMAP) were used to reduce the number of highly variable genes.
Cell-cell interaction analysis
The R package “CellChat” (https://github.com/sqjin/CellChat) was utilized to analyze and visualize the intercellular communication among different cell types and specific ligand-receptor interactions based on single-cell RNA sequencing (scRNA-seq) data.
Public data sources and acquisition
Single-cell RNA-seq data from Salcher et al. were downloaded from the core atlas of Single-Cell Lung Cancer Atlas (LuCA) (https://cellxgene.cziscience.com/collections/edb893ee-4066-4128-9aec-5eb2b03f8287) (31). This large integrated Seurat object comprising data for 156 LUAD patients and 86 control individuals was utilized to reveal distinct TME and cell compositions. The gene expression data of the TCGA-LUAD and GSE72094 cohorts were normalized by the “Scale” function before analysis. Missing value from bulk mRNA data were imputated by mean value, and missing clinical data was deleted. The protein expression levels of NFRGs were compared using the online website The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) (http://ualcan.path.uab.edu/analysis.html).
Construction of an NFRG-based prognostic signature
Twenty-four NFRGs were included from previous studies, and they are presented in Table S1 (32-35). The R package “limma” (version 3.36.2) was used to identify differentially expressed NFRGs between LUAD tumor tissues and noncancer tissue samples. Benjamini-Hochberg (BH) method from the “limma” package to control the false discovery rate (FDR) when identifying differential genes, taking into account that performing multiple hypothesis tests may lead to false-positive results. The univariate Cox proportional hazards regression model was performed to identify genes with a significant prognostic value. Furthermore, the LASSO Cox regression model via the R package “glmnet” (version 4.1-2) was used to inform the selection of the most prognostic gene signature from all the identified NFRGs and obviate model overfitting. The optimal tuning parameter λ was identified via the 1-SE (standard error) criterion. According to the risk scores derived from the selected gene signature, a prognostic model was developed. The risk score was the sum of the messenger RNA (mRNA) expression levels of the four NFRGs and their corresponding coefficients, i.e., risk score = ∑[CAT*(−0.220535782175529) + CTSG*(−0.0368657515179532) + ENO1*(0.156333581719548) + TLR2*(−0.150335127077594)].
Evaluation of the prognostic performance of the NFRG-based signature
A LUAD cohort (GSE72094) was employed as the test cohort. Patients were divided into high- and low-risk groups based on the median risk score. Kaplan-Meier (K-M) survival curves and PCA were used to assess the predictive performance of the signature. Receiver operating characteristic (ROC) curves were used to compare the accuracy of 1-, 3- and 5-year survival predictions. In addition, univariate and multivariate Cox regression models were utilized to evaluate the clinical variables (age, sex and clinical stage) of patients in the training and testing cohorts, in combination with the risk score generated by our prognostic model. The “prcomp” function in the R package “stats” was used to perform PCA. The R packages “survival”, “survminer”, “timeROC” and “survex” were used for ROC curve analysis and Brier score evaluation.
Construction of a nomogram for survival prediction
A clinically applicable nomogram based on prognostic variables (age, sex, clinical stage and risk score) of the pooled cohort was constructed to predict 1-, 3-, and 5-year survival probabilities. The predictive performance of the nomogram was validated through ROC curve analysis.
Single-sample gene set enrichment analysis (ssGSEA) evaluation of the top 10 oncogenic pathways
The top 10 oncogenic pathways, involving 330 reported genes, were selected to evaluate the enrichment score using the ssGSEA algorithm (36). Representative scores were calculated as the activated score minus the repressed score as previously reported (37). Subsequently, we compared the ssGSEA score of each pathway between distinct risk groups. The risk groups were determined based on the risk score associated with the newly established prognostic signature.
Potential chemotherapy therapeutic drug sensitivity prediction
The “pRRophetic” package was used to predict the difference in drug sensitivity between the high- and low-risk groups (38).
Characterization of the tumor immune microenvironment
The R package “gsva” was utilized to perform ssGSEA to calculate the scores of 15 infiltrating immune cells and to evaluate the activity of 14 immune-related pathways. The R package “ESTIMATE” was used to predict tumor purity as well as the proportion of stromal and immune cells in the TME (39).
Immunotherapeutic response prediction
The machine learning tool Tumor Immune Dysfunction and Exclusion (TIDE) is applied to predict cancer patients’ response to immune checkpoint inhibitors (ICIs) (40). The TIDE score, T-cell exclusion score, myeloid-derived suppressor cell (MDSC) score and cancer-associated fibroblast (CAF) score were retrieved from the TIDE portal (http://tide.dfci.harvard.edu) on the basis of normalized transcriptome data for the TCGA-LUAD cohort. Moreover, the Wilcoxon rank-sum test was conducted to compare the mRNA levels of immune checkpoint molecules in the TCGA-LUAD cohort between the low- and high-risk groups. Furthermore, the GSE126044 and GSE135222 cohorts, comprising patients with NSCLC who received anti-programmed cell death ligand 1 (anti-PD-L1) or anti-programmed cell death protein 1 (anti-PD-1) antibody therapy, were used as external cohorts (41,42). The risk scores between responders (R) and non-responders (NR) in the above cohorts were compared.
Evaluation of the prognostic signature by quantitative real-time polymerase chain reaction (PCR)
Fifty-five pairs of LUAD and matched adjacent normal tissue samples were collected for reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Samples were obtained from patients who had been diagnosed with primary LUAD by pathological examination of tissue biopsy and had surgical resection operations at the Cancer Hospital of the Chinese Academy of Medical Sciences (Beijing, China) from September 2021 to March 2022. The clinical information for all patients enrolled is listed in Table S2. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Peking Union Medical College Cancer Hospital (Beijing, China) (No. NCC2021C-527). Written informed consent was obtained from all participants.
Tissue specimens were immediately stored in liquid nitrogen after resection. Total RNA was extracted from the tissue specimens using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. SuperScript II reverse transcriptase was used to synthesize first-strand cDNA from the RNA template (TaKaRa, Japan, RR047). Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing) was used to perform RT-qPCR. The relative expression levels were normalized to those of glyceraldehyde phosphate dehydrogenase (GAPDH). Table S3 shows the sequences of the primers utilized in our study. Patients were divided into two groups based on the calculated risk score.
Statistical analysis
Cox prediction models were applied to evaluate the sample size. The K-M method with a two-sided log-rank test was performed to compare the overall survival (OS) of patients between subgroups. To assess the independent prognostic value of the risk model, we used univariate and multivariate Cox regression models. The Wilcoxon rank sum test was used to assess the immune cell infiltration and immune pathways between the two groups. The Wilcoxon rank-sum test or Kruskal-Wallis test was performed to analyze continuous variables. All statistical analyses were accomplished with R software (version 4.1.2), SPSS 22.0 (IBM Corp., Armonk, New York, USA) and GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, CA, USA). A P value lower than 0.05 was considered statistically significant.
Results
Analysis of the TME and NFRGs at single-cell resolution
To investigate the involvement of NETs in LUAD, we utilized single-cell RNA sequencing (scRNA-seq) data from 156 LUAD patients and 86 control individuals to reveal distinct TMEs and cell compositions. A total of 623,816 cells were included (Figure 1A). Overall, 24 cell types were identified in total (Figure 1B). Compared to normal tissues, LUAD tissues exhibited the highest proportions of CD4 T cells and CD8 T cells. In addition, tumor cells, regulatory T cells (Tregs) and neutrophils were overwhelmingly increased in LUAD tissues (Figure 1C). To further elucidate the interactions between the above cells, cell-cell communication analysis was conducted (Figure 1D). The potential outgoing and incoming signals as well as the paired molecules in specific signaling pathways were assessed. As illustrated in Figure 1D, tumor cells were the major signal provider and receptor. Neutrophils appeared to send signals by the secreted class 3 semaphorin protein (SEMA3) signaling pathway and receive signals by the noncanonical WNT (ncWNT) signaling pathway (Figure 1E). The role of neutrophils in promoting tumors is partly due to their ability to produce NETs (43). Subsequently, we sought to determine the differential expression of NFRGs among distinct cell types. According to Figure 1F, TLR2, TNF, S100A8, S100A9, MMP9, LYZ, Enolase 1 (ENO1), CTSC, CAT, CTSG, BPI, and AZU1 were expressed in almost every cell type. In contrast, TLR2, S100A12, and CTSG were specifically expressed in neutrophils. In addition, TKT, LTF and AZU1 were mostly expressed in T cells, ciliated cells and myeloid cells (Figure 1F). Overall, these findings suggest that NETs play a key role in the development of LUAD.

Identification of NFRGs between LUAD tumor tissue and normal tissue samples and construction of a prognostic signature
To assess the differentially expressed NFRGs, data for 497 cancer and 54 normal tissue samples were downloaded from the TCGA database. A heatmap illustrated that 21 NFRGs were differentially expressed between tumor and adjacent nontumor tissues (Figure 2A). To construct a prognostic model, univariate Cox regression analysis was performed to screen genes with a prognostic value. As a result, six genes (CAT, CTSG, ENO1, MNDA, TLR2 and TLR4) met the criteria of P<0.05 and thus were considered related to survival. Among them, ENO1 was associated with an increased risk of mortality with a hazard ratio (HR) >1, while the other five genes (CAT, CTSG, MNDA, TLR2 and TLR4) were protective genes with HRs <1 (Figure 2B). Furthermore, the protein levels of these 6 genes were compared using the online website UALCAN. CAT, CTSG, and MNDA were downregulated, while ENO1 and TLR2 were upregulated in primary tumor tissues. However, the protein expression level of TLR4 seemed to be extremely low (Figure 2C). Next, LASSO Cox regression analysis was performed to construct a 4-gene-based signature according to the optimum λ value (Figure 2D,2E). The risk score was calculated as follows: risk score = ∑[CAT*(−0.220535782175529) + CTSG*(−0.0368657515179532) + ENO1*(0.156333581719548) + TLR2*(−0.150335127077594)] (Figure 2F).

Evaluation of the prognostic performance of the NFRG-based signature
To validate the prognostic model, TCGA-LUAD patients were employed as our training cohort, and a GEO cohort containing 393 LUAD patients (GSE72094) was employed as the test cohort. Patients were divided into high- and low-risk groups based on the median risk score. K-M analysis indicated a significant difference in the survival rate between the low- and high-risk groups (P<0.001, Figure 3A,3B). Patients in the low-risk group (blue dotted line) were found to have longer survival times and lower death rates than those in the high-risk group (red dotted line). In addition, ROC curve analysis showed that our signature showed robust predictive efficacy in the training cohort (AUC =0.677 for 1-year, 0.639 for 3-year, and 0.583 for 5-year survival) (Figure 3C) and test cohort (AUC =0.653 for 1-year, 0.673 for 3-year, and 0.747 for 5-year survival) (Figure 3D). Risk plots exhibited satisfactory separation between the two groups (Figure 3E,3F). Additionally, Brier score in both train cohort and test cohort were less than 0.25, which suggested a robust predicting power of the signature (Figure 3G,3H).

Independent prognostic value of the risk signature and construction of a nomogram for survival prediction
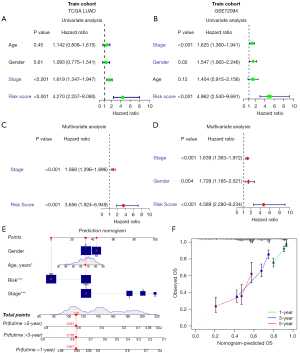
Univariate and multivariate Cox regression analyses were conducted to evaluate whether the risk signature could independently predict prognosis. Univariate Cox regression analysis showed that our risk signature was an independent prognostic factor in the TCGA (HR =4.270, 95% CI: 2.257–8.080, Figure 4A) and GEO cohorts (HR: 4.962, 95% CI: 2.540–9.691, Figure 4B). Multivariate analysis indicated that the risk score was still a prognostic factor in both cohorts after adjusting for other confounding factors (HR =3.656, 95% CI: 1.924–6.949, Figure 4C; HR =4.589, 95% CI: 2.280–9.234, Figure 4D). Additionally, to develop a more applicable quantitative analytic algorithm that could be used in the clinic to predict LUAD patients’ expected survival, a nomogram was constructed by integrating the risk status and common clinical characteristics (age, sex, clinical stage, and risk score). These variables were employed to calculate the individual sample’s score as well as the total score for evaluating 1-, 3- and 5-year survival probabilities (Figure 4E). Moreover, calibration curves were developed to show the consistency between the actual prognostic value and the value calculated by the nomogram. The calibration curves exhibited a near optimal performance, suggesting that our predictive nomogram was accurate (Figure 4F).
Correlations between the risk signature and clinicopathological characteristics
Based on the above, we found a significant independent prognostic risk signature for patients with LUAD. To further assess the roles of NFRGs in the development of LUAD, we sought to explore clinicopathological features that were associated with the risk signature. All patients in the TCGA-LUAD cohort were ranked by the formula score and then divided into high- and low-risk groups. Age, the extent and size of the primary tumor, and distant metastasis status had no significant correlation with the risk score (Figure 5A). In contrast, patients who had higher risk scores seemed to have an advanced clinical stage (Figure 5B) and more severe lymph node metastases (Figure 5C). Interestingly, except for Epidermal Growth Factor Receptor (EGFR) mutation, the relative risk scores were obviously higher for the group with other mutation statuses compared with the wild-type group (Figure 5D). In addition, patients with Tumor Protein p53 (TP53) mutations had the highest risk. Furthermore, we performed a stratified survival analysis and identified that higher risk scores were associated with unfavorable clinical outcomes only in the KRAS proto-oncogene, GTPase (KRAS) mutation group, not in TP53 and EGFR co-mutation group and wild type group (Figure 5E-5G). These results indicate that NFRGs are closely associated with LUAD progression and prognosis.

Relationship between the prognostic risk signature and chemotherapy drug sensitivity
Functional annotation was further performed using ssGSEA. Based on previously reported signatures (Table S4), we calculated the enrichment scores of the top 10 common oncogenic pathways in the high- and low-risk groups. The MYC, NOTCH, nuclear factor erythroid 2-related factor 2 (NRF2), Wnt/β-catenin (WNT), phosphatidylinositol 3-kinase (PI3K) and cell cycle–related pathways had higher ssGSEA scores in the high-risk group; conversely, the receptor tyrosine kinase/RAS (RTK/RAS) pathway was remarkably enriched in the low-risk group (Figure 6A). Accordingly, we evaluated the sensitivity of chemotherapeutic drugs targeting different oncogenic pathways. Patients in the low-risk group exhibited lower half-maximal inhibitory concentration (IC50) values for drugs targeting the cell cycle (Figure 6B), PI3K (Figure 6C), WNT (Figure 6D), c-Jun N-terminal kinase (JNK) (Figure 6E), and poly ADP-ribose polymerase (PARP) (Figure 6F) pathways. These results suggested that the risk signature helped to predict chemotherapy drug sensitivity in LUAD patients.

Assessment of the TME and prediction of immunotherapy response
To evaluate the distinct TME between the low- and high-risk groups, we further compared the enrichment scores of 16 types of immune cells and the activity of 15 infiltrating immune cells and 14 immune-related pathways in the TCGA cohort by applying the ssGSEA score. A heatmap illustrated that antitumor immune cells were excessively enriched in the low-risk group, suggesting that the low-risk group exhibited a more active immune microenvironment (Figure 7A). In addition, the Estimation of STromal and Immune cells in MAlignant Tumor tissues (ESTIMATE) algorithm was used to quantify the overall infiltrating immune cells and stromal cells. The results confirmed that the low-risk patients had higher immune scores and ESTIMATE scores than the high-risk individuals in the TCGA cohort (Figure 7B).

Given the above results, we hypothesized that patients in the low-risk group might gain more benefit from immunotherapies. It has been reported that TIDE is an accurate method for predicting cancer patient responsiveness to ICI therapies (40). As expected, patients in the high-risk group had higher TIDE scores, T-cell exclusion scores, CAF scores and MDSC scores, indicating a higher probability of tumor immune escape and a lower likelihood of benefiting from anti-PD-1/CTLA4 therapy (Figure 7C). Moreover, the mRNA expression levels of CTLA4, TIGIT, HAVCR2 and CD47 were remarkably upregulated in the low-risk group (Figure 7D). Moreover, risk scores were slightly higher in non-responders (NR) than in responders (R) in two independent NSCLC immune checkpoint blockade (ICB) cohorts. Altogether, we concluded that the risk signature could serve as a biomarker for predicting immunotherapeutic benefits.
Evaluation of the expression levels of the four NFRGs in LUAD tumor tissues by RT-qPCR
We subsequently validated our bioinformatics analysis results in our own cohort of 58 LUAD patients. RT-qPCR was utilized to quantify the expression levels of the four NFRGs (CAT, CTSG, ENO1, TLR2). Compared with the corresponding nontumor tissue samples, the relative mRNA expression levels of ENO1 and TLR2 were significantly higher, but those of CTSG and CAT were lower in tumor tissue samples (Figure 8A). Based on our risk formula, every patient received a risk score, so they were divided into low- and high-risk groups based on the median risk score. Patients who had lymph node metastasis and distant metastasis had higher risk scores (Figure 8B,8C). Similarly, the lower the differential status was, the higher the risk score (Figure 8D). Moreover, the ratio plot showed that patients in the high-risk group had more severe lymph node metastasis (Figure 8E) and distant metastasis (Figure 8F) and worse differential status (Figure 8G).

Discussion
In this study, we first evaluated the composition of the TME in LUAD patients at single-cell resolution. Similar to Treg cells, tumor-associated neutrophils were substantially increased in LUAD samples. Previous studies have indicated that neutrophils are the most dominant immune cells in NSCLC and might have multiple cross-talk connections with other cells (44-48). Our analysis illustrated that neutrophils mainly interacted with tumor cells through the SEMA3 and ncWNT signaling pathways, which are associated with tumor angiogenesis, malignant cell proliferation, invasiveness, and metastasis (49-51). More importantly, the tumor-promoting role of neutrophils is in part conferred by their ability to produce NETs (43). NET formation occurs when neutrophils are stimulated by microorganisms, cytokines, chemicals, immune complexes or tumor cells (52-54).
Therefore, we characterized the differential expression of NFRGs at both the scRNA-seq and bulk RNA-seq levels in LUAD samples. We found that most of these genes were significantly differentially expressed; more importantly, six genes (CAT, CTSG, ENO1, TLR2, TLR4, MNDA) were considered independent prognostic factors.
Based on these survival-related genes, we constructed a 4-gene risk signature through LASSO Cox regression analysis. Its prognostic value was then validated to be robust and sensitive in training and test cohorts. Moreover, as chemotherapy is one of the main treatment strategies for LUAD, we systematically explored the discrepancy in chemotherapy drug sensitivity between the low-risk and high-risk groups. Notably, patients in the low-risk group exhibited lower IC50 values for drugs that target the cell cycle, PI3K, WNT, JNK and PARP pathways. Taken together, our risk model might provide new insight for the individual management of LUAD patients with distinct risk scores.
Further analysis indicated that LUAD patients who had an advanced clinical stage and severe lymph node metastasis tended to have higher risk scores. Interestingly, the role that NETs play in tumor metastasis is an emerging topic of interest. On the one hand, NETs have been shown to promote tumor cell migration and invasion in LUAD (11). On the other hand, NET accumulation in the lungs leads to dormant cell awakening and subsequent metastatic growth (55). Strikingly, our stratified survival analysis indicated that higher risk scores were associated with unfavorable clinical outcomes specifically in the TP53 and EGFR co-mutation group and KRAS mutation group, which are the most commonly mutated genes in LUAD (1). A previous study has suggested that KRAS-mutant tumor cells may induce stronger NETs of tumor-infiltrating neutrophils through upregulation of CXCL8 (56). To a certain extent, our risk model helped to stratify LUAD patients with distinct clinical and mutational statuses. In addition, NETs might serve as a potential target for LUAD patients with KRAS mutations.
Currently, immunotherapy has exhibited great potential as a novel strategy for LUAD treatment (57). However, responses to immunotherapy vary among individuals, possibly due to the low tumor mutational burden (TMB), immunosuppressive TME, and low expression of immune checkpoint molecules (58,59). Therefore, discovering effective biomarkers for predicting which groups of LUAD patients could benefit from immunotherapy is of great urgency. Our study demonstrated that antitumor immune cells were substantially enriched and that the ESTIMATE score was significantly higher in the low-risk group, indicating a more active immune microenvironment. Moreover, our 4-NFRG-based prognostic risk score was negatively correlated with the expression levels of immune checkpoint molecules such as CTLA4, TIGIT, HAVCR2 and CD47, indicating that the 4-NFRG-based prognostic model may play a pivotal role in predicting the immunotherapy response. In addition, patients who had higher risk scores also had higher TIDE scores, T-cell exclusion scores, CAF scores and MDSC scores, indicating a higher probability of tumor immune escape and a lower likelihood of benefiting from anti-PD-1/CTLA4 therapy. Furthermore, two NSCLC immunotherapy cohorts, GSE126044 and GSE135222, were applied to evaluate the efficacy of our risk score for predicting the response to immunotherapy. In summary, our 4-NFRG-based prognostic signature is an effective biomarker for predicting immunotherapeutic benefits.
Next, the clinical relevance of our risk score was validated in a clinical cohort consisting of 58 LUAD patients. The relative mRNA expression levels of the four NFRGs (CAT, CTSG, ENO1, TLR2) were quantified by RT-qPCR. Consistent with the bioinformatics analysis, the expression levels of ENO1 and TLR2 were significantly higher, while those of CTSG and CAT were lower in tumor samples than in their corresponding nontumor samples. In addition, based on the risk formula generated in our study, the risk scores for the 58 patients were determined, and the patients were divided into low- and high-risk groups according to the median risk score. As expected, we found that patients with higher risk scores tended to have worse differential status and more severe lymph node metastasis.
Nonetheless, there are still several limitations in our study. First, for every LUAD patient, the relationship between the 4-NFRG-based risk score and the real amounts of NETs in tumor tissues is unclear. In the future, efforts should be made to evaluate their correlation and to explore more representative markers of NET formation. Moreover, the NSCLC immunotherapy cohorts enrolled in this study were relatively small. We believe that the predictive value of our risk score for immunotherapy response needs to be adjusted and explored in multicenter and large-scale prospective cohorts.
Conclusions
In brief, we systematically evaluated NFRGs at both the single-cell and bulk mRNA levels. We comprehensively validated the predictive value of the novel four NFRGs-based risk model on the prognosis of LUAD patients in the TCGA cohort, GEO cohort and clinical samples. Our risk signature helped to stratify LUAD patients with distinct clinical and mutational statuses. Furthermore, our 4-NFRGs-based risk signature may be an effective biomarker to predict the immunotherapy response in LUAD patients. Finally, validating NETs formation might promote individual risk stratification, and targeting NETs might offer personalized therapy strategies for patients with LUAD.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-463/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-463/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-463/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-463/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Peking Union Medical College Cancer Hospital (Beijing, China) (No. NCC2021C-527). Written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Riely GJ, Wood DE, Ettinger DS, et al. Non-Small Cell Lung Cancer, Version 4.2024, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2024;22:249-74. [Crossref] [PubMed]
- Zilionis R, Engblom C, Pfirschke C, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 2019;50:1317-1334.e10. [Crossref] [PubMed]
- Chen J, Tan Y, Sun F, et al. Single-cell transcriptome and antigen-immunoglobin analysis reveals the diversity of B cells in non-small cell lung cancer. Genome Biol 2020;21:152. [Crossref] [PubMed]
- Kim N, Kim HK, Lee K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020;11:2285. [Crossref] [PubMed]
- Leader AM, Grout JA, Maier BB, et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 2021;39:1594-1609.e12. [Crossref] [PubMed]
- Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol 2019;16:601-20. [Crossref] [PubMed]
- Burgener SS, Schroder K. Neutrophil Extracellular Traps in Host Defense. Cold Spring Harb Perspect Biol 2020;12:a037028. [Crossref] [PubMed]
- Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science 2004;303:1532-5. [Crossref] [PubMed]
- Mousset A, Bellone L, Gaggioli C, et al. NETscape or NEThance: tailoring anti-cancer therapy. Trends Cancer 2024;10:655-67. [Crossref] [PubMed]
- Cools-Lartigue J, Spicer J, McDonald B, et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest 2013;1233446-58. [Crossref] [PubMed]
- Yazdani HO, Roy E, Comerci AJ, et al. Neutrophil Extracellular Traps Drive Mitochondrial Homeostasis in Tumors to Augment Growth. Cancer Res 2019;79:5626-39. [Crossref] [PubMed]
- Houghton AM, Rzymkiewicz DM, Ji H, et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med 2010;16:219-23. [Crossref] [PubMed]
- Rayes RF, Mouhanna JG, Nicolau I, et al. Primary tumors induce neutrophil extracellular traps with targetable metastasis promoting effects. JCI Insight 2019;5:e128008. [Crossref] [PubMed]
- Xiao Y, Cong M, Li J, et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell 2021;39:423-437.e7. [Crossref] [PubMed]
- Rayes J, Brill A. Hot under the clot: venous thrombogenesis is an inflammatory process. Blood 2024;144:477-89. [Crossref] [PubMed]
- Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol 2012;198:773-83. [Crossref] [PubMed]
- Zhang Y, Hu Y, Ma C, et al. Diagnostic, Therapeutic Predictive, and Prognostic Value of Neutrophil Extracellular Traps in Patients With Gastric Adenocarcinoma. Front Oncol 2020;10:1036. [Crossref] [PubMed]
- Rivera-Franco MM, Leon-Rodriguez E, Torres-Ruiz JJ, et al. Neutrophil Extracellular Traps Associate with Clinical Stages in Breast Cancer. Pathol Oncol Res 2020;26:1781-5. [Crossref] [PubMed]
- Rosell A, Aguilera K, Hisada Y, et al. Prognostic value of circulating markers of neutrophil activation, neutrophil extracellular traps, coagulation and fibrinolysis in patients with terminal cancer. Sci Rep 2021;11:5074. [Crossref] [PubMed]
- Chen X, Ma H, Mo S, et al. Intratumoral neutrophil extracellular traps are associated with unfavorable clinical outcomes and immunogenic context in pancreatic ductal adenocarcinoma. Front Immunol 2022;13:1027459. [Crossref] [PubMed]
- Chen J, Wang T, Li X, et al. DNA of neutrophil extracellular traps promote NF-κB-dependent autoimmunity via cGAS/TLR9 in chronic obstructive pulmonary disease. Signal Transduct Target Ther 2024;9:163. [Crossref] [PubMed]
- de Andrea CE, Ochoa MC, Villalba-Esparza M, et al. Heterogenous presence of neutrophil extracellular traps in human solid tumours is partially dependent on IL-8. J Pathol 2021;255:190-201. [Crossref] [PubMed]
- Kaltenmeier C, Yazdani HO, Morder K, et al. Neutrophil Extracellular Traps Promote T Cell Exhaustion in the Tumor Microenvironment. Front Immunol 2021;12:785222. [Crossref] [PubMed]
- Vesely MD, Zhang T, Chen L. Resistance Mechanisms to Anti-PD Cancer Immunotherapy. Annu Rev Immunol 2022;40:45-74. [Crossref] [PubMed]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity 2009;30:646-55. [Crossref] [PubMed]
- Saravia J, Chapman NM, Chi H. Helper T cell differentiation. Cell Mol Immunol 2019;16:634-43. [Crossref] [PubMed]
- Basu A, Ramamoorthi G, Albert G, et al. Differentiation and Regulation of T(H) Cells: A Balancing Act for Cancer Immunotherapy. Front Immunol 2021;12:669474. [Crossref] [PubMed]
- Cheng Y, Gong Y, Chen X, et al. Injectable adhesive hemostatic gel with tumor acidity neutralizer and neutrophil extracellular traps lyase for enhancing adoptive NK cell therapy prevents post-resection recurrence of hepatocellular carcinoma. Biomaterials 2022;284:121506. [Crossref] [PubMed]
- Egan K, Cooke N, Kenny D. Living in shear: platelets protect cancer cells from shear induced damage. Clin Exp Metastasis 2014;31:697-704. [Crossref] [PubMed]
- Salcher S, Sturm G, Horvath L, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell 2022;40:1503-1520.e8. [Crossref] [PubMed]
- Shen XT, Xie SZ, Xu J, et al. Pan-Cancer Analysis Reveals a Distinct Neutrophil Extracellular Trap-Associated Regulatory Pattern. Front Immunol 2022;13:798022. [Crossref] [PubMed]
- Zhang Y, Guo L, Dai Q, et al. A signature for pan-cancer prognosis based on neutrophil extracellular traps. J Immunother Cancer 2022;10:e004210. [Crossref] [PubMed]
- Wang H, Zhang Y, Wang Q, et al. The regulatory mechanism of neutrophil extracellular traps in cancer biological behavior. Cell Biosci 2021;11:193. [Crossref] [PubMed]
- Vorobjeva NV, Pinegin BV. Neutrophil extracellular traps: mechanisms of formation and role in health and disease. Biochemistry (Mosc) 2014;79:1286-96. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-337.e10. [Crossref] [PubMed]
- Xiao Y, Ma D, Zhao S, et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin Cancer Res 2019;25:5002-14. [Crossref] [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468. [Crossref] [PubMed]
- Tamborero D, Rubio-Perez C, Muiños F, et al. A Pan-cancer Landscape of Interactions between Solid Tumors and Infiltrating Immune Cell Populations. Clin Cancer Res 2018;24:3717-28. [Crossref] [PubMed]
- Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550-8. [Crossref] [PubMed]
- Cho JW, Hong MH, Ha SJ, et al. Genome-wide identification of differentially methylated promoters and enhancers associated with response to anti-PD-1 therapy in non-small cell lung cancer. Exp Mol Med 2020;52:1550-63. [Crossref] [PubMed]
- Jung H, Kim HS, Kim JY, et al. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat Commun 2019;10:4278. [Crossref] [PubMed]
- Masucci MT, Minopoli M, Del Vecchio S, et al. The Emerging Role of Neutrophil Extracellular Traps (NETs) in Tumor Progression and Metastasis. Front Immunol 2020;11:1749. [Crossref] [PubMed]
- Stankovic B, Bjørhovde HAK, Skarshaug R, et al. Immune Cell Composition in Human Non-small Cell Lung Cancer. Front Immunol 2018;9:3101. [Crossref] [PubMed]
- Kargl J, Busch SE, Yang GH, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun 2017;8:14381. [Crossref] [PubMed]
- Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest 2014;124:5466-80. [Crossref] [PubMed]
- Yang WC, Hwang YS, Chen YY, et al. Interleukin-4 Supports the Suppressive Immune Responses Elicited by Regulatory T Cells. Front Immunol 2017;8:1508. [Crossref] [PubMed]
- Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer 2016;16:431-46. [Crossref] [PubMed]
- Toledano S, Nir-Zvi I, Engelman R, et al. Class-3 Semaphorins and Their Receptors: Potent Multifunctional Modulators of Tumor Progression. Int J Mol Sci 2019;20:556. [Crossref] [PubMed]
- Tamagnone L. Emerging role of semaphorins as major regulatory signals and potential therapeutic targets in cancer. Cancer Cell 2012;22:145-52. [Crossref] [PubMed]
- Lee EH, Chari R, Lam A, et al. Disruption of the non-canonical WNT pathway in lung squamous cell carcinoma. Clin Med Oncol 2008;2008:169-79.
- Clark SR, Ma AC, Tavener SA, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med 2007;13:463-9. [Crossref] [PubMed]
- Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J Exp Med 2020;217:e20200652. [Crossref] [PubMed]
- Yang L, Liu Q, Zhang X, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 2020;583:133-8. [Crossref] [PubMed]
- Albrengues J, Shields MA, Ng D, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 2018;361:eaao4227. [Crossref] [PubMed]
- Shang A, Gu C, Zhou C, et al. Exosomal KRAS mutation promotes the formation of tumor-associated neutrophil extracellular traps and causes deterioration of colorectal cancer by inducing IL-8 expression. Cell Commun Signal 2020;18:52. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Negrao MV, Skoulidis F, Montesion M, et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J Immunother Cancer 2021;9:e002891. [Crossref] [PubMed]
- Shao XM, Huang J, Niknafs N, et al. HLA class II immunogenic mutation burden predicts response to immune checkpoint blockade. Ann Oncol 2022;33:728-38. [Crossref] [PubMed]


