
Prevalence of EGFR gene mutations in patients with early-stage resectable non-small cell lung cancer in Spain: the ORIGEN study
Highlight box
Key findings
• Epidermal growth factor receptor (EGFR) mutations were detected by IdyllaTM EGFR Mutation Test, a polymerase chain reaction-based molecular test, in 14.5% of patients.
• Next-generation sequencing (NGS) provided consistent results but detected a higher percentage of EGFR mutations and additional actionable drivers and concurrent genomic alterations.
What is known and what is new?
• Studies of early-stage non-small cell lung cancer (NSCLC) in Western populations have shown EGFR mutation rates similar to those in advanced-stage NSCLC patients.
• The prevalence of EGFR mutations in patients with early-stage resected NSCLC has not been previously reported in Spain.
What is the implication, and what should change now?
• Molecular testing is crucial in early-stage NSCLC and can be performed either with single-gene testing or NGS.
Introduction
Epidermal growth factor receptor (EGFR) mutations are the second most common oncogenic driver in non-small cell lung cancer (NSCLC), following KRAS mutations (1,2). Exon 19 deletions and the exon 21 L858R point mutations are the most frequent EGFR actionable alterations, accounting for 85–90% of all EGFR mutations. Less frequent sensitizing EGFR alterations include exon 20 insertions (4–12%) and the following single nucleotide variants (SNVs): G719X, S768I, and L861Q (3%) (2-6).
The standard of care for patients with advanced NSCLC harboring actionable common EGFR mutations is first-line osimertinib (7). In surgically resected tumors harboring common EGFR mutations, the ADAURA study demonstrated that adjuvant osimertinib provided a significant benefit in terms of disease-free survival (DFS) (8) and overall survival (OS) (9). This study established adjuvant osimertinib therapy as a new standard of care for these patients. Therefore, the detection of EGFR mutations in surgically resected NSCLC is crucial for selecting patients who are candidates to adjuvant osimertinib (3). Molecular testing to identify EGFR mutations is recommended by guidelines not only for advanced NSCLC, but also for early-stage NSCLC (10-16).
Previous studies reported that the frequency of EGFR mutations in advanced NSCLC ranges from 12.8% to 14.1% in Europe (3,4). Consistently, in Spain, several observational studies conducted between 2005 and 2015 identified sensitizing EGFR mutations in 11.6% to 16.6% of patients with advanced NSCLC (17-19). Studies on early-stage NSCLC in Western populations have reported EGFR mutation rates comparable to those observed in advanced NSCLC patients (20-22).
To our knowledge, the frequency of EGFR mutations in patients with early stage resected NSCLC in Spain has not been previously investigated. This study provides information about the prevalence of this genomic alteration in Spain, but also contributes to understanding the variability in EGFR mutation rates in early-stage NSCLC across European countries.
We conducted a multicenter, observational study to determine the prevalence of EGFR mutations in patients with surgically resected early stage (IA to IIIB) non-squamous NSCLC in Spain using real-time polymerase chain reaction (PCR)-based molecular EGFR mutation tests. We used this PCR-based technique because in access to NGS in Spain is not universal, especially in the context of early-stage disease. To complement this information, we also evaluated the utility of next-generation sequencing (NGS) in the same cohort, as some centers had already adopted this approach and to compare the results obtained using both methods. We present this article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1146/rc).
Methods
Study design
This observational, non-interventional, multicenter, cross-sectional study was conducted at 19 centers in Spain (Table S1). Preliminary results from this study were presented at the ESMO Congress 2023 (23). The study was performed in accordance with the applicable local regulations for non-interventional and/or observational studies and following the ethical principles of the Declaration of Helsinki (as revised in 2013) and Good Clinical Practice guidelines (24). The study protocol and informed consent forms were reviewed and approved by the Ethical Committee of the University Hospital 12 de Octubre (Madrid, Spain; No. CEIm 21/230, 25 May 2021). All the study participants provided informed consent before undergoing any study procedure.
Study population
Patients were eligible for the study if they were male or female, were older than 18 years of age, had been histologically or cytologically diagnosed with early-stage [i.e., IA to IIIB, American Joint Committee on Cancer (AJCC) 8th Edition] (25) non-squamous NSCLC, and had their tumor surgically removed within 6 weeks before enrollment, or had undergone planned surgery within 8 weeks after inclusion in the study. Patients who presented with metastatic or unresectable tumors were excluded.
Study objectives
The primary endpoint of the study was to determine the prevalence of sensitizing EGFR mutations using real-time PCR-based molecular analysis in patients with early-stage nonsquamous NSCLC with stage IA–IIIB disease (according to the AJCC 8th), who underwent surgical resection in Spain. The secondary endpoints were to describe the baseline characteristics of the study population, to compare the results obtained using the IdyllaTM EGFR Mutation Test (Biocartis, Mechelen, Belgium) with those obtained using the Oncomine™ Precision Assay GX (Thermo Fisher Scientific Inc., Waltham, MA, USA) and to determine the prevalence of uncommon EGFR mutations.
Study assessments and data collection
Data on the following variables were collected during the study visit from the available medical records: demographics, characteristics of the tumor [e.g., histological type, clinical and pathological staging according to the AJCC 8th Edition, type of diagnostic biopsy, adjuvant therapy, and time from first visit to any medical consultation (primary care or specialist care) until surgery], presence of residual tumor (R0 or R1), and EGFR mutation results based on the IdyllaTM EGFR Mutation Test and Oncomine™ Precision Assay. Both tests are described in Table S2, and the EGFR mutations that were detected by the IdyllaTM EGFR Mutation Test are shown in Table S3. The presence of concurrent pathogenic genomic alterations was assessed using the Oncomine™ Precision Assay. Compared to the IdyllaTM EGFR Mutation Test, NGS performed with the Oncomine™ Precision Assay can identify distinct EGFR mutations in exons 18–21. Furthermore, the Oncomine™ Precision Assay is capable of simultaneously detecting hotspot mutations, copy number variations, and gene fusions involving 50 key genes, such as EGFR, ALK, ROS1, RET, NTRK and KRAS.
Surgical samples were analyzed at the local laboratory of each participating center using the IdyllaTM EGFR Mutation Test, which is able to detect 51 EGFR mutations including exon 18 (G719A/S/C), 36 deletions in exon 19, 2 exon 20 mutations (T790M, S768I), 5 insertions in exon 20 (c.2310_2311insGGT; p.D770_N771insG and c.2319_2320insCAC; p.H773_V774insH) and 2 exon 21 mutations (L858R, L861Q). However, it does not detect all EGFR exon 20 insertion mutations, as some variants fall outside its coverage (26,27). The Oncomine™ Precision Assay was performed on all surgical samples from the study subjects who provided their informed consent for NGS testing. The analysis was conducted in two central laboratories (Molecular Biology Core Facility from the Hospital Clínic of Barcelona and the Laboratori Core d’Anàlisi Molecular from the Hospital Universitari of Bellvitge and Institut Català d’Oncologia, Barcelona; Spain).
Statistical analysis
Based on the number of newly diagnosed NSCLC patients (n=22,000) in Spain in 2020 (28) and assuming that the percentage of patients with early stage and surgically resected NSCLC is approximately 25%, it was estimated that a total sample of 173 patients with early stage and surgically resected NSCLC would be required to detect a prevalence of EGFR mutations of 11.7%, with a precision of ±5%, and assuming a dropout rate of 10%.
The statistical analyses were primarily descriptive. Categorical variables are presented as absolute and relative frequencies. Continuous variables are presented using descriptive statistics (mean and standard deviation or median and interquartile range). Missing data were not imputed.
The prevalence of sensitizing EGFR mutations in the study population was based on the percentage of evaluable patients with common sensitizing EGFR mutations (deletion of exon 19 and point mutation in exon 21), as detected by the IdyllaTM EGFR Mutation Test and the Oncomine™ Precision Assay; these results are presented with the corresponding 95% confidence intervals (CIs). The concordance between the IdyllaTM EGFR Mutation Test and the Oncomine Precision Assay in surgical samples was assessed using Cohen’s kappa index. Additionally, the sensitivity, specificity, and positive and negative predictive values of the IdyllaTM EGFR Mutation Test for comparing the mutation status between diagnostic samples (bronchoscopy/thoracic puncture) and surgical samples were calculated for the evaluable study population.
Finally, a multivariate logistic regression model was used to estimate the associations between EGFR mutation status and demographic and clinical characteristics of the evaluable study population. The following variables were included in the model: EGFR mutation status (EGFR mutated versus EGFR wild-type) of tumor samples as the dependent variable and key demographic and clinical characteristics, such as age (continuous), sex (female vs. male), histology (adenocarcinoma vs. non-adenocarcinoma), programmed death-ligand 1 (PD-L1) status (positive vs. negative) and Eastern Cooperative Oncology Group (ECOG) performance status (0 vs. ≥1), as independent variables. A stepwise backward approach was used for fitting the regression model.
All the analyses were performed using IBM SPSS version 26 software.
Results
Patients’ disposition and characteristics
Between August 2021 and February 2022, 183 patients were consecutively enrolled in the study by the Departments of Oncology of 19 Spanish centers. Of these, 172 met all the selection criteria and were ultimately included in the analyses (Figure S1). The patients had a median age of 67.5 years, 57.6% of the patients were male, and 83.1% were current or former smokers. The most common histological type was adenocarcinoma (96.5%), and most patients included in the study had stage IA/IB disease (65.1%). The baseline clinical characteristics of the patients are shown in Table 1. Additionally, only 38 (22.9%) of 166 patients with available information planned to receive adjuvant chemotherapy. The adjuvant chemotherapy regimens according to disease stage are shown in Table S4.
Table 1
| Characteristics | Total (n=172) | EGFR wild type (n=147) | EGFR mutated (n=25) |
|---|---|---|---|
| Age (years) | 67.5 (61.0–72.5) | 68.0 (61.0–72.0) | 66.0 (61.0–73.0) |
| Sex | |||
| Male | 99 (57.6) | 93 (63.3) | 6 (24.0) |
| Female | 73 (42.4) | 54 (36.7) | 19 (76.0) |
| Smoking history | |||
| Smoker | 54 (31.4) | 50 (34.0) | 4 (16.0) |
| Former smoker | 89 (51.7) | 80 (54.4) | 9 (36.0) |
| Non-smoker | 24 (14.0) | 14 (9.5) | 10 (40.0) |
| Passive smoker | 1 (0.6) | 0 (0.0) | 1 (4.0) |
| Unknown | 4 (2.3) | 3 (2.0) | 1 (4.0) |
| Histology | |||
| Adenocarcinoma | 166 (96.5) | 141 (95.9) | 25 (100.0) |
| Large cell carcinoma | 3 (1.7) | 3 (2.0) | 0 (0.0) |
| Sarcomatoid carcinoma | 1 (0.6) | 1 (0.7) | 0 (0.0) |
| Carcinoid tumor | 1 (0.6) | 1 (0.7) | 0 (0.0) |
| Adenosquamous carcinoma | 1 (0.6) | 1 (0.7) | 0 (0.0) |
| Pathological stage | |||
| IA | 81 (47.1) | 71 (48.3) | 10 (40.0) |
| IB | 31 (18.0) | 24 (16.3) | 7 (28.0) |
| IIA | 7 (4.1) | 7 (4.8) | 0 (0.0) |
| IIB | 25 (14.5) | 19 (12.9) | 6 (24.0) |
| IIIA | 19 (11.0) | 17 (11.6) | 2 (8.0) |
| IIIB | 4 (2.3) | 4 (2.7) | 0 (0.0) |
| Unknown | 5 (2.9) | 5 (3.4) | 0 (0.0) |
| Diagnostic sample | |||
| Biopsy | 16 (9.3) | 15 (10.2) | 1 (4.0) |
| Bronchoscopy | 30 (17.4) | 23 (15.6) | 7 (28.0) |
| Intraoperative diagnosis | 1 (0.6) | 1 (0.7) | 0 (0.0) |
| Surgery | 91 (52.9) | 74 (50.3) | 17 (68.0) |
| Trans-thoracic needle biopsy | 34 (19.8) | 34 (23.1) | 0 (0.0) |
| Surgery | |||
| Lobectomy | 135 (78.9) | 115 (78.8) | 20 (80.0) |
| Segmentectomy | 25 (14.6) | 22 (15.1) | 3 (12.0) |
| Pneumonectomy | 5 (2.9) | 4 (2.7) | 1 (4.0) |
| Wedge resection | 6 (3.5) | 5 (3.4) | 1 (4.0) |
| Adjuvant chemotherapy | |||
| No | 128 (77.1) | 110 (78.0) | 18 (72.0) |
| Yes | 38 (22.9) | 31 (22.0) | 7 (28.0) |
Data are presented as median (IQR) or n (%). There are missing data in the following characteristics: ECOG PS, surgery, resection margin and adjuvant chemotherapy. ECOG, European Cooperative Oncology Group; EGFR, epidermal growth factor receptor; IQR, interquartile range; PS, performance status; R0, complete tumor resection; R1, microscopic residual tumor.
Frequency of EGFR mutations as determined by the IdyllaTM EGFR Mutation Test
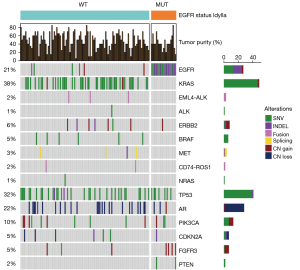
EGFR mutations were detected in 25 patients (14.5%; 95% CI: 0.7–28.3%) using the IdyllaTM EGFR Mutation Test. Subjects harboring sensitizing EGFR mutations were more likely to be females (76% vs. 36.7%) and nonsmokers (40% vs. 9.5%) (Table 1). The following EGFR mutations were detected in this study using the IdyllaTM EGFR Mutation Test: exon 19 deletion in 13 patients (7.6%), exon 21 L858R point mutation in 11 patients (6.4%), and exon 20 mutation (T790M) in 1 patient (0.6%) (Figure 1). The list of EGFR mutations detected using the IdyllaTM test and clinical staging are shown in Table 2.
Table 2
| Stage | Mutated | Exon 19 deletion | L858R mutation | Exon 20 mutation |
|---|---|---|---|---|
| IA (n=81) | 10 (12.3) | 4 (4.9) | 6 (7.4) | 0 (0.0) |
| IB (n=31) | 7 (22.6) | 4 (12.9) | 3 (9.7) | 0 (0.0) |
| IIA (n=7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| IIB (n=25) | 6 (24.0) | 3 (12.0) | 2 (8.0) | 1 (4.0) |
| IIIA (n=19) | 2 (10.5) | 2 (10.5) | 0 (0.0) | 0 (0.0) |
| IIIB (n=4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Unknown (n=5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Total (n=172) | 25 (14.5) | 13 (7.6) | 11 (6.4) | 1 (0.6) |
Data are presented as n (%). EGFR, epidermal growth factor receptor; TNM, tumor, node, metastasis.
Frequency of EGFR mutations as determined by the OncomineTM Precision Assay
The OncomineTM Precision Assay was performed on surgical samples from 128 patients. Compared to the IdyllaTM test results shown above, EGFR mutations were detected in 25 (19.5%) of the 128 patients (Table 3). More specifically, the OncomineTM test detected exon 19 deletions in 10 patients (7.8%), exon 21 mutations in 10 patients (7.8%), and exon 20 insertions in 5 patients (3.9%). The following exon 20 mutations were identified using the OncomineTM test: c.2319_2320insTGTCCACAC, c.2314_2319dup, c.2308G>A, c.2408G>T, and c.2308_2309insCCAGCGTGG. Interestingly, the IdyllaTM and OncomineTM tests detected different mutations in exon 20 in the same patient, namely, T790M (c.2369C>T) and c.2308_2309insCCAGCGTGG, respectively.
Table 3
| Variable | IdyllaTM | OncomineTM Precision Assay |
|---|---|---|
| Samples tested | 172 | 128* |
| EGFR mutation | 25 (14.5) | 25 (19.5) |
| Other EGFR alteration | – | 1 amplification |
| Exon 19 deletion | 13 (52.0) | 10 (40.0) |
| Exon 20 mutation | 1 (4.0) | 5 (20.0) |
| L858R mutation | 11 (44.0) | 10 (40.0) |
Data are presented as n (%). *, the OncomineTM Precision Assay was not performed in four patients with EGFR mutations detected by IdyllaTM. Among these patients, three patients had exon 19 deletions, and one had an exon 21 L858R mutation. EGFR, epidermal growth factor receptor.
The concordance analysis between the two tests showed a kappa coefficient of 0.89 (Table S5). Furthermore, using OncomineTM as a reference test, the IdyllaTM test showed high specificity (100%), sensitivity (84.0%; 95% CI: 77.6–90.4%), positive predictive value (100%), and negative predictive value (96.3%; 95% CI: 93.0–99.5%). A subsequent concordance analysis between the two tests excluding the EGFR exon 20 insertion mutations yielded a kappa coefficient of 1.00.
We performed a multivariate logistic regression analysis, and we observed that age [odds ratio (OR) 0.973, 95% CI: 0.964–0.983] and female sex (OR 2.435, 95% CI: 1.058–5.604) were significantly associated with EGFR mutation detected by the OncomineTM Precision Assay (Table S6).
Additional alterations identified by OncomineTM Precision Assay
The OncomineTM Precision Assay enabled the detection of concurrent genomic alterations in patients with EGFR-mutant NSCLC (Figure 2), being the most common concurrent alterations in these patients TP53, FGFR3, PIK3CA, PTEN, and CDKN2A. Furthermore, NGS allowed the identification of additional actionable alterations in the remaining patients: KRASG12C (22%), ERBB2 (6%), METex14 (2%), EML4-ALK fusions (2%), BRAFV600E (2%), and CD74-ROS1 fusions (2%). We also detected ALK imbalance in 6 cases (4%) and 1 case with RET imbalance (1%). An ALK SNV (C1156*) was detected in 1 patient (variant allele frequency =0.038).
Discussion
In our study, we found that 14.5% of the patients harbored a sensitizing EGFR mutation. This frequency is consistent with the results of other studies conducted in Western countries that included patients with early-stage NSCLC. Similarly, as observed in other studies, patients with EGFR-mutated tumors in our study were more likely to be female, nonsmokers, and have adenocarcinoma histology (20,21,29,30). EGFR testing using the IdyllaTM test showed that the most frequent variants consisted of exon 19 deletions, followed by exon 21 L858R point mutations, accounting for more than 90% of all the sensitizing mutations that were detected. Exon 20 mutations were found in only 4% of patients. Similar rates have been previously reported in similar studies assessing the frequency of EGFR mutations in surgical samples from early-stage NSCLC patients (20,21,29,30). To our knowledge, this is the first epidemiological study to assess the prevalence of sensitizing EGFR mutations in patients with surgically resected early-stage NSCLC in Spain.
In our study, we used two EGFR testing methods: the IdyllaTM EGFR Mutation Test, which is a fully automated, real-time (RT) quantitative polymerase chain reaction (qPCR)-based molecular test that covers 51 mutations in exons 18–21, and the OncomineTM precision assay, which is an integrated NGS-based test. The OncomineTM precision assay is considered to be a more sensitive method than qPCR because it covers the entire EGFR coding sequence and allows the assessment of additional concurrent genomic alterations, covering 50 cancer driver genes. Both tests provided consistent results regarding exon 19 and exon 21 mutations, but the OncomineTM test detected a higher percentage of EGFR mutations, with five additional mutations located in exon 20 that were not covered by the IdyllaTM test (31). Moreover, by using the OncomineTM test, we detected additional concurrent genomic alterations in patients with EGFR-mutant NSCLC, such as TP53, FGFR3, PIK3CA, PTEN, and CDKN2A, as well as additional actionable alterations in patients with wild-type EGFR, such as KRAS G12C, BRAF V600E and ERBB2 mutations, METex14 and ALK and ROS1 fusions. These findings are clinically relevant because the presence of concurrent mutations in TP53 and PIK3CA has been associated with a worse prognosis and a greater risk of histological transformation upon osimertinib treatment (32-35). Furthermore, we believe that this information may be highly relevant for clinical practice, considering that new additional targeted therapies, such as alectinib, an ALK inhibitor, have demonstrated clinical benefit in the adjuvant setting for ALK rearranged tumors, as observed in the ALINA study (36). In addition, detecting other actionable alterations, such as RET rearrangements, could allow the enrollment of patients in ongoing clinical trials. On the other hand, the incorporation of immunotherapy in the adjuvant setting is also increasing the relevance of molecular testing in surgically resected NSCLC, as EGFR- and ALK-positive tumors are not deemed candidates for adjuvant atezolizumab or pembrolizumab. In fact, real-world evidence and guidelines endorse the use of NGS rather than RT-PCR as the preferred method for identifying a wider array of actionable EGFR mutations in NSCLC (12,16,26,37). However, by the time the present study was conducted, access to NGS testing for EGFR mutation screening and detection had not been universally implemented within the Spanish Health System. However, although the OncomineTM test is a more sensitive technique for detecting EGFR mutations, allowing the detection of some false negatives that are missed by conventional testing methods, the IdyllaTM test is simple, fast, widely implemented, and reliable for detecting common EGFR mutations. Furthermore, both techniques showed high concordance, and the IdyllaTM test showed high sensitivity, specificity, and predictive value. A similar high concordance between RT-PCR and NGS testing technologies has been previously reported (27).
The emergence of EGFR tyrosine kinase inhibitors (EGFR-TKIs) has changed the paradigm for the management of EGFR-mutated NSCLC. Several EGFR mutations are sensitive to TKIs, particularly osimertinib, a third-generation EGFR-TKI that prolongs progression-free survival and OS in advanced and early-stage EGFR-mutated NSCLC patients (8,9,38). However, none of the patients included in the study received adjuvant osimertinib because it was not reimbursed in Spain when the study was conducted. Currently, osimertinib is reimbursed by the Spanish National Health System for patients with completely resected stage IB-IIIA NSCLC with common EGFR mutations. In addition, although it is not yet a widely adopted standard practice, reflex single gene or NGS testing has been recommended to optimize the molecular characterization of NSCLC (27). A process in which the pathologist is responsible for initiating and controlling testing for a set of preapproved biomarkers (including EGFR) at the time of initial diagnosis, without direct oncologist involvement, has contributed to enhancing the quality of biomarker testing, shortening turnaround time, and improving patient outcomes (11,37,39-41). Moreover, recently published international guidelines recommend reflex biomarker testing for all patients diagnosed with non-squamous NSCLC, regardless of disease stage (16).
Our study had several limitations. Despite consecutive and prospective sampling, some sources of selection bias cannot be ruled out. We included only patients with non-squamous NSCLC, as EGFR mutations are generally more clinically relevant and prevalent in this histological subtype (8). In addition, NGS was not conducted for all patients because not all patients provided the consent or have adequate material.
Conclusions
In conclusion, the prevalence of EGFR mutations in early stage, resectable, nonsquamous NSCLC in Spain is consistent with the observed frequency in advanced NSCLC and with previous reports in this clinical setting. Whenever feasible, NGS is the technology of choice because it provides a more complete genomic profile. However, single-gene testing could also be considered a valid method to screen for common EGFR mutations in early-stage NSCLC to identify patients who are candidates for adjuvant osimertinib, following the recommendations of the Spanish and European clinical guidelines (12,15).
Acknowledgments
The authors thank María Isabel López, Susana Vara, Andrea Barchino, Ana Moreno, Juan Luis Sanz and Fernando Rico-Villademoros (APICES, Madrid; Spain) for their support with the study setup, coordination and project management, monitoring, statistical analysis and editorial assistance. Preliminary results from this study were presented at the ESMO Congress 2023.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1146/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1146/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1146/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1146/coif). C.T. reports grants or contracts and consulting fees from Novartis; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from AstraZeneca, MSD, Genotipia, Biocartis, Janssen; support for attending meetings and/or travel from MSD, Pfizer, Lilly; participation on a Data Safety Monitoring Board or Advisory Board from MSD, AstraZeneca; she is a member of the board of Societat Catalana de Citopatologia. C.A.F. reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Pfizer, AstraZeneca and Bayer; patents planned, issued or pending from Roche, Ipsen and Pfizer. H.A. reports grants or contracts from Asociación Española Contra el Cancer and Ferrer Farma; payment or honoraria as speaker from Takeda; support for attending meetings and/or travel from Roche, BMS, MSD, and Takeda. M.L. reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from MSD, AstraZéneca, BMS, Ipsen, Roche, Lilly and Bayer; payment for expert testimony from MSD, BMS and Merck; support for attending meetings and/or travel from Ipsen, MSD and Roche. V.C. reports consulting fees from Roche, Bristol Myers Squibb, Merck Sharp & Dohme, AstraZeneca, Takeda, Pfizer, Sanofi and AMGEN; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Roche, Bristol Myers Squibb/Celgene, Merck Sharp & Dohme, AstraZeneca, Takeda Sanofi and AMGEN; support for attending meetings and/or travel from Takeda, Roche, Bristol Myers Squibb and Merck Sharp & Dohme. R.A. reports consulting fees from PHARMAMAR, NOVARTIS, ROCHE, ASTRAZENECA; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from DECIPHERA, BOEHRINGER; support for attending meetings and/or travel from PHARMAMAR, ROCHE, MSD; participation on a Data Safety Monitoring Board or Advisory Board from NOVARTIS. J.B. reports grants or contracts from any entity to the institution from SEOM; consulting fees from Astra Zeneca, Roche, BMS; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Lilly; payment for expert testimony from Roche; support for attending meetings and/or travel from MSD and Johnson and Johnson. J.V. reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Sanofi; support for attending meetings and/or travel from Roche, MSD, Pfizer, Pierre Fabre, Amgen; participation on a Data Safety Monitoring Board or Advisory Board from Sanofi. E.A. reports consulting fees from Boehringer-Ingelheim, Lilly, Pfizer, Roche; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from AstraZeneca, MSD, TAKEDA, Roche, Lilly, Pfizer; support for attending meetings and/or travel from Astra Zeneca, Roche; has been Steering committee member of the NEOLA trial, LIBRETTO-431, EVOLVE- LUNG 02. R.B. reports investigational grant from ROCHE; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from ROCHE, BMS, PFIZER, MSD, AMGEN, TAKEDA, ASTRAZENECA; participation on a Data Safety Monitoring Board or Advisory Board from TAKEDA, ROCHE, BMS, ASTRA ZENECA. D.I. reports consulting fees from ROCHE, ASTRAZENECA, BMS, MSD, JOHNSON & JOHNSON, PHARMAMAR, PFIZER; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events PFIZER, JOHNSON & JOHNSON, BMS, MSD; support for attending meetings and/or travel from MSD, ASTRAZENECA, ROCHE. C.C. reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Roche, Lilly, AstraZeneca, Pfizer; support for attending meetings and/or travel from Lilly, Roche, DAKO/AGILENT. B.M. reports personal fees, non-financial support and other from Roche, personal fees and other from MSD, personal fees and other from Astra Zeneca, personal fees and other from Bristol Myers Squibb, personal fees from Takeda, outside the submitted work. A.B. reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Takeda, Regeneron, GSK, Roche, Astra Zeneca; support for attending meetings and/or travel Daiichi Sankyo, Roche, Takeda. T.G. reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from MSD and Bristol Myers; support for attending meetings and/or travel from MSD; she is treasurer of the Sociedad Andaluza de Oncologia Médica. Manuel Cobo reports Consulting fees from Novartis, AstraZeneca, Boehringer-Ingelheim, Roche, BMS, Lilly, MSD, Takeda, Phyzer, Kyowa, Sanofi, Jansen; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Novartis, AstraZeneca, Boehringer-Ingelheim, Roche, BMS, Lilly, MSD, Takeda, Kyowa, Pierre-fabre, Novocure, Sanofi, Jansen. Marc Campayo reports consulting fees from Astra Zeneca, Boehringer Ingelheim; payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Astra Zeneca, EUSA Pharma, Ipsen, Lilly, Merck, Sharp & Dohme, Novartis, Pfizer, Roche, Sanofi-Aventis, Takeda; support for attending meetings and/or travel AstraZeneca, Lilly, Merck, Sharp & Dohme, Pfizer, Roche, Sanofi-Aventis. A.C. reports to be an employee of AstraZeneca Farmacéutica Spain. M.D. reports to be AstraZeneca Farmacéutica Spain employee. E.N. reports research grants from Roche, Pfizer, BMS and Merck Serono; consulting fees from Roche, Bristol Myers Squibb, Merck Sharp Dohme, Merck-Serono, Sanofi, Pfizer, Lilly, Amgen, Janssen, Daiichi-Sankyo, Boehringer-Ingelheim, AstraZeneca, Takeda, Sanofi, Janssen, Pierre Fabre and Qiagen; honoraria for lectures from Roche, Bristol Myers Squibb, Merck Sharp Dohme, Sanofi, Pfizer, Lilly, Amgen, Janssen, Boehringer-Ingelheim, AstraZeneca, Takeda, Sanofi, Janssen and Qiagen; support for attending meetings and/or travel from Takeda, MSD, Roche, Pfizer; participation on a Data Safety Monitoring Board or Advisory Board of Roche, Apollomics, Transgene and Daiichi. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was performed in accordance with the applicable local regulations for noninterventional and/or observational studies and following the ethical principles of the Declaration of Helsinki (as revised in 2013) and Good Clinical Practice guidelines. The study protocol and informed consent forms were reviewed and approved by the Ethical Committee of the University Hospital 12 de Octubre (Madrid, Spain; No. CEIm 21/230, 25 May 2021). All the study participants provided informed consent before undergoing any study procedure.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chevallier M, Borgeaud M, Addeo A, et al. Oncogenic driver mutations in non-small cell lung cancer: Past, present and future. World J Clin Oncol 2021;12:217-37. [Crossref] [PubMed]
- Harrison PT, Vyse S, Huang PH. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol 2020;61:167-79. [Crossref] [PubMed]
- Melosky B, Kambartel K, Häntschel M, et al. Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis. Mol Diagn Ther 2022;26:7-18. [Crossref] [PubMed]
- Zhang YL, Yuan JQ, Wang KF, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016;7:78985-93. [Crossref] [PubMed]
- John T, Taylor A, Wang H, et al. Uncommon EGFR mutations in non-small-cell lung cancer: A systematic literature review of prevalence and clinical outcomes. Cancer Epidemiol 2022;76:102080. [Crossref] [PubMed]
- Russano M, Perrone G, Di Fazio GR, et al. Uncommon EGFR mutations in non-small-cell lung cancer. Precis Cancer Med 2022;5:30. [Crossref]
- Greenhalgh J, Boland A, Bates V, et al. First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer. Cochrane Database Syst Rev 2021;3:CD010383. [PubMed]
- Wu YL, Tsuboi M, He J, et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:1711-23. [Crossref] [PubMed]
- Tsuboi M, Herbst RS, John T, et al. Overall Survival with Osimertinib in Resected EGFR-Mutated NSCLC. N Engl J Med 2023;389:137-47. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2022;20:497-530. [Crossref] [PubMed]
- Gosney JR, Paz-Ares L, Jänne P, et al. Pathologist-initiated reflex testing for biomarkers in non-small-cell lung cancer: expert consensus on the rationale and considerations for implementation. ESMO Open 2023;8:101587. [Crossref] [PubMed]
- Isla D, Lozano MD, Paz-Ares L, et al. New update to the guidelines on testing predictive biomarkers in non-small-cell lung cancer: a National Consensus of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin Transl Oncol 2023;25:1252-67. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. [Crossref] [PubMed]
- Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-iv21. [Crossref] [PubMed]
- Remon J, Soria JC, Peters S, et al. Early and locally advanced non-small-cell lung cancer: an update of the ESMO Clinical Practice Guidelines focusing on diagnosis, staging, systemic and local therapy. Ann Oncol 2021;32:1637-42. [Crossref] [PubMed]
- Passaro A, Leighl N, Blackhall F, et al. ESMO expert consensus statements on the management of EGFR mutant non-small-cell lung cancer. Ann Oncol 2022;33:466-87. [Crossref] [PubMed]
- Cortes-Funes H, Gomez C, Rosell R, et al. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann Oncol 2005;16:1081-6. [Crossref] [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [Crossref] [PubMed]
- Esteban E, Majem M, Martinez Aguillo M, et al. Prevalence of EGFR mutations in newly diagnosed locally advanced or metastatic non-small cell lung cancer Spanish patients and its association with histological subtypes and clinical features: The Spanish REASON study. Cancer Epidemiol 2015;39:291-7. [Crossref] [PubMed]
- Hondelink LM, Ernst SM, Atmodimedjo P, et al. Prevalence, clinical and molecular characteristics of early stage EGFR-mutated lung cancer in a real-life West-European cohort: Implications for adjuvant therapy. Eur J Cancer 2023;181:53-61. [Crossref] [PubMed]
- Sara Kuruvilla M, Liu G, Syed I, et al. EGFR mutation prevalence, real-world treatment patterns, and outcomes among patients with resected, early-stage, non-small cell lung cancer in Canada. Lung Cancer 2022;173:58-66. [Crossref] [PubMed]
- Mordant P MD. Outcome of Patients With Resected Early-Stage Non-small Cell Lung Cancer and EGFR Mutations: Results From the IFCT Biomarkers France Study. Clin Lung Cancer 2023;24:1-10. [Crossref] [PubMed]
- Nadal E, Fernandez CA, Arasanz H, et al. 1278P ORIGEN: Multicenter study on the prevalence of EGFR gene mutations in patients with early-stage resectable non-small cell lung cancer in Spain. Ann Oncol 2023;34:S738-9. [Crossref]
- World Medical Association Inc. DECLARATION OF HELSINKI: ethical principles for medical research involving human subjects. 2008. Available online: https://www.wma.net/wp-content/uploads/2016/11/DoH-Oct2008.pdf
- Lababede O, Meziane MA. The Eighth Edition of TNM Staging of Lung Cancer: Reference Chart and Diagrams. Oncologist 2018;23:844-8.
- Pisapia P, Russo A, De Luca C, et al. The relevance of the reference range for EGFR testing in non-small cell lung cancer patients. Lung Cancer 2024;198:108002. [Crossref] [PubMed]
- Goffinet S, Bontoux C, Heeke S, et al. EGFR status assessment using reflex testing targeted next-generation sequencing for resected non-squamous non-small cell lung cancer. Virchows Arch 2025;486:531-9. [Crossref] [PubMed]
- Sociedad Española de Oncología Médica. Las cifras del cáncer en España. 2020. Available online: https://seom.org/seomcms/images/stories/recursos/Cifras_del_cancer_2020.pdf
- Soo RA, Reungwetwattana T, Perroud HA, et al. Prevalence of EGFR Mutations in Patients With Resected Stages I to III NSCLC: Results From the EARLY-EGFR Study. J Thorac Oncol 2024;19:1449-59. [Crossref] [PubMed]
- Batra U, Prabhash K, Noronha V, et al. Prevalence of EGFR Mutations in Patients With Resected Stage I to III Nonsquamous Non-Small Cell Lung Cancer: Results of India Cohort. JCO Glob Oncol 2025;11:e2400353. [Crossref] [PubMed]
- Pacini L, Jenks AD, Vyse S, et al. Tackling Drug Resistance in EGFR Exon 20 Insertion Mutant Lung Cancer. Pharmgenomics Pers Med 2021;14:301-17. [Crossref] [PubMed]
- Aggarwal C, Davis CW, Mick R, et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non-Small-Cell Lung Cancer. JCO Precis Oncol 2018;2018:PO.18.00107.
- Lan B, Zhao N, Du K, et al. Concurrent TP53 mutations predict a poor prognosis of EGFR-mutant NSCLCs treated with TKIs: An updated systematic review and meta-analysis. Oncol Lett 2022;24:384. [Crossref] [PubMed]
- Qin K, Hou H, Liang Y, et al. Prognostic value of TP53 concurrent mutations for EGFR- TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: a meta-analysis. BMC Cancer 2020;20:328. [Crossref] [PubMed]
- Qiu X, Wang Y, Liu F, et al. Survival and prognosis analyses of concurrent PIK3CA mutations in EGFR mutant non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors. Am J Cancer Res 2021;11:3189-200. [PubMed]
- Solomon BJ, Ahn JS, Dziadziuszko R, et al. LBA2 ALINA: efficacy and safety of adjuvant alectinib versus chemotherapy in patients with early-stage ALK+ non-small cell lung cancer (NSCLC). Ann Oncol 2023;34:S1295-6. [Crossref]
- Hofman P, Calabrese F, Kern I, et al. Real-world EGFR testing practices for non-small-cell lung cancer by thoracic pathology laboratories across Europe. ESMO Open 2023;8:101628. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Anand K, Phung TL, Bernicker EH, et al. Clinical Utility of Reflex Ordered Testing for Molecular Biomarkers in Lung Adenocarcinoma. Clin Lung Cancer 2020;21:437-42. [Crossref] [PubMed]
- Gregg JP, Li T, Yoneda KY. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey. Transl Lung Cancer Res 2019;8:286-301. [Crossref] [PubMed]
- Pasello G, Lorenzi M, Pretelli G, et al. Diagnostic-Therapeutic Pathway and Outcomes of Early Stage NSCLC: a Focus on EGFR Testing in the Real-World. Front Oncol 2022;12:909064. [Crossref] [PubMed]



